This is a preprint.
Efficient and multiplexed tracking of single cells using whole-body PET/CT
- PMID: 37662335
- PMCID: PMC10473747
- DOI: 10.1101/2023.08.23.554536
Efficient and multiplexed tracking of single cells using whole-body PET/CT
Update in
-
Ultrasensitive and multiplexed tracking of single cells using whole-body PET/CT.Sci Adv. 2024 Jun 14;10(24):eadk5747. doi: 10.1126/sciadv.adk5747. Epub 2024 Jun 14. Sci Adv. 2024. PMID: 38875333 Free PMC article.
Abstract
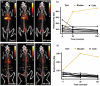
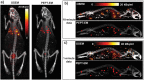
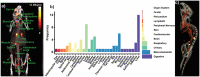
In vivo molecular imaging tools are crucially important for elucidating how cells move through complex biological systems, however, achieving single-cell sensitivity over the entire body remains challenging. Here, we report a highly sensitive and multiplexed approach for tracking upwards of 20 single cells simultaneously in the same subject using positron emission tomography (PET). The method relies on a new tracking algorithm (PEPT-EM) to push the cellular detection threshold to below 4 Bq/cell, and a streamlined workflow to reliably label single cells with over 50 Bq/cell of 18F-fluorodeoxyglucose (FDG). To demonstrate the potential of method, we tracked the fate of over 70 melanoma cells after intracardiac injection and found they primarily arrested in the small capillaries of the pulmonary, musculoskeletal, and digestive organ systems. This study bolsters the evolving potential of PET in offering unmatched insights into the earliest phases of cell trafficking in physiological and pathological processes and in cell-based therapies.
Keywords: FDG; cell tracking; positron emission tomography; reconstruction algorithms; single-cell analysis.
Figures



References
-
- Hong H., Yang Y., Zhang Y. & Cai W. Non-Invasive Cell Tracking in Cancer and Cancer Therapy. Current Topics in Medicinal Chemistry 10, 1237–1248 (2010). https://doi.org/http://dx.doi.org.stanford.idm.oclc.org/10.2174/156802610791384234 - DOI - PMC - PubMed
-
- Lauri C., Varani M., Bentivoglio V., Capriotti G. & Signore A. Present status and future trends in molecular imaging of lymphocytes. Seminars in Nuclear Medicine 53, 125–134 (2023). https://doi.org/10.1053/j.semnuclmed.2022.08.011 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
