This is a preprint.
Deep screening of proximal and distal splicing-regulatory elements in a native sequence context
- PMID: 37662340
- PMCID: PMC10473672
- DOI: 10.1101/2023.08.21.554109
Deep screening of proximal and distal splicing-regulatory elements in a native sequence context
Update in
-
CRISPR-dCas13d-based deep screening of proximal and distal splicing-regulatory elements.Nat Commun. 2024 May 7;15(1):3839. doi: 10.1038/s41467-024-47140-8. Nat Commun. 2024. PMID: 38714659 Free PMC article.
Abstract
Pre-mRNA splicing, a key process in gene expression, can be therapeutically modulated using various drug modalities, including antisense oligonucleotides (ASOs). However, determining promising targets is impeded by the challenge of systematically mapping splicing-regulatory elements (SREs) in their native sequence context. Here, we use the catalytically dead CRISPR-RfxCas13d RNA-targeting system (dCas13d/gRNA) as a programmable platform to bind SREs and modulate splicing by competing against endogenous splicing factors. SpliceRUSH, a high-throughput screening method, was developed to map SREs in any gene of interest using a lentivirus gRNA library that tiles the genetic region, including distal intronic sequences. When applied to SMN2, a therapeutic target for spinal muscular atrophy, SpliceRUSH robustly identified not only known SREs, but also a novel distal intronic splicing enhancer, which can be targeted to alter exon 7 splicing using either dCas13d/gRNA or ASOs. This technology enables a deeper understanding of splicing regulation with applications for RNA-based drug discovery.
Keywords: SMN2; alternative splicing; antisense oligonucleotides (ASO); dCas13d/guide RNA; high-throughput screen; splicing-regulatory elements (SREs).
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT Y.R., D.U., and C.Z. are inventors on a patent application submitted based on this work. C.Z. is a co-founder of a startup company working on ASO therapeutics. Y.T.Y., X.W., M.J., L.V.Y., and Q.W. declare no competing interests.
Figures
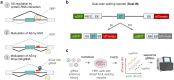

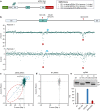
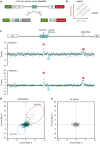
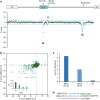
References
-
- Gilbert W. (1978). Why genes in pieces? Nature 271, 501–501. - PubMed
-
- Chow L.T., Gelinas R.E., Broker T.R., and Roberts R.J. (1977). An amazing sequence arrangement at the 5’ ends of adenovirus 2 messenger RNA. Cell 12, 1–8. - PubMed
-
- Pan Q., Shai O., Lee L.J., Frey B.J., and Blencowe B.J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genet 40, 1413–1415. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
