This is a preprint.
Antibodies Expand the Scope of Angiotensin Receptor Pharmacology
- PMID: 37662341
- PMCID: PMC10473732
- DOI: 10.1101/2023.08.23.554128
Antibodies Expand the Scope of Angiotensin Receptor Pharmacology
Update in
-
Antibodies expand the scope of angiotensin receptor pharmacology.Nat Chem Biol. 2024 Dec;20(12):1577-1585. doi: 10.1038/s41589-024-01620-6. Epub 2024 May 14. Nat Chem Biol. 2024. PMID: 38744986 Free PMC article.
Abstract
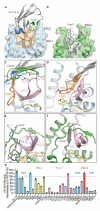
G protein-coupled receptors (GPCRs) are key regulators of human physiology and are the targets of many small molecule research compounds and therapeutic drugs. While most of these ligands bind to their target GPCR with high affinity, selectivity is often limited at the receptor, tissue, and cellular level. Antibodies have the potential to address these limitations but their properties as GPCR ligands remain poorly characterized. Here, using protein engineering, pharmacological assays, and structural studies, we develop maternally selective heavy chain-only antibody ("nanobody") antagonists against the angiotensin II type I receptor (AT1R) and uncover the unusual molecular basis of their receptor antagonism. We further show that our nanobodies can simultaneously bind to AT1R with specific small-molecule antagonists and demonstrate that ligand selectivity can be readily tuned. Our work illustrates that antibody fragments can exhibit rich and evolvable pharmacology, attesting to their potential as next-generation GPCR modulators.
Conflict of interest statement
Competing Interests A.C.K., C.M., L.W., D.P.S., and M.A.S., are co-inventors on patent application for AT1R blocking nanobodies. A.C.K. is a cofounder and consultant for biotechnology companies Tectonic Therapeutic and Seismic Therapeutic, and also for the Institute for Protein Innovation, a nonprofit research institute. L.M.W. is a scientific advisor for Septerna. D.P.S. is a Septerna employee. C.M. is a Sanofi employee.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
