This is a preprint.
Inhibition of JAK-STAT pathway corrects salivary gland inflammation and interferon driven immune activation in Sjögren's Disease
- PMID: 37662351
- PMCID: PMC10473773
- DOI: 10.1101/2023.08.16.23294130
Inhibition of JAK-STAT pathway corrects salivary gland inflammation and interferon driven immune activation in Sjögren's Disease
Update in
-
Inhibition of JAK-STAT pathway corrects salivary gland inflammation and interferon driven immune activation in Sjögren's disease.Ann Rheum Dis. 2024 Jul 15;83(8):1034-1047. doi: 10.1136/ard-2023-224842. Ann Rheum Dis. 2024. PMID: 38527764 Free PMC article.
Abstract
Objectives: Inflammatory cytokines that signal through the JAK- STAT pathway, especially interferons (IFNs), are implicated in Sjögren's Disease (SjD). Although inhibition of JAKs is effective in other autoimmune diseases, a systematic investigation of IFN-JAK-STAT signaling and effect of JAK inhibitor (JAKi) therapy in SjD-affected human tissues has not been reported.
Methods: Human minor salivary glands (MSGs) and peripheral blood mononuclear cells (PBMCs) were investigated using bulk or single cell (sc) RNA sequencing (RNAseq), immunofluorescence microscopy (IF), and flow cytometry. Ex vivo culture assays on PBMCs and primary salivary gland epithelial cell (pSGEC) lines were performed to model changes in target tissues before and after JAKi.
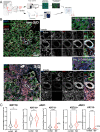
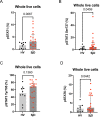
Results: RNAseq and IF showed activated JAK-STAT pathway in SjD MSGs. Elevated IFN-stimulated gene (ISGs) expression associated with clinical variables (e.g., focus scores, anti-SSA positivity). scRNAseq of MSGs exhibited cell-type specific upregulation of JAK-STAT and ISGs; PBMCs showed similar trends, including markedly upregulated ISGs in monocytes. Ex vivo studies showed elevated basal pSTAT levels in SjD MSGs and PBMCs that were corrected with JAKi. SjD-derived pSGECs exhibited higher basal ISG expressions and exaggerated responses to IFNβ, which were normalized by JAKi without cytotoxicity.
Conclusions: SjD patients' tissues exhibit increased expression of ISGs and activation of the JAK-STAT pathway in a cell type-dependent manner. JAKi normalizes this aberrant signaling at the tissue level and in PBMCs, suggesting a putative viable therapy for SjD, targeting both glandular and extraglandular symptoms. Predicated on these data, a Phase Ib/IIa randomized controlled trial to treat SjD with tofacitinib was initiated.
Keywords: Interferon; Janus kinases; STAT; Sjögren’s Disease; Tofacitinib.
Conflict of interest statement
Potential Conflicts of Interest: BMW has Cooperative Research Award and Development Agreements [CRADA] from Pfizer, Inc., and Mitobridge, Inc. (A subsidiary of Astellas Pharma, Inc.). NIAMS has CRADAs with Astra Zeneca and Bristol Myers Squibb. These CRADA did not financially support the experimental results presented herein.
Figures







References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
