This is a preprint.
Spatial lipidomics of fresh-frozen spines
- PMID: 37662353
- PMCID: PMC10473750
- DOI: 10.1101/2023.08.23.554488
Spatial lipidomics of fresh-frozen spines
Abstract
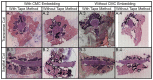
Technologies assessing the lipidomics, genomics, epigenomics, transcriptomics, and proteomics of tissue samples at single-cell resolution have deepened our understanding of physiology and pathophysiology at an unprecedented level of detail. However, the study of single-cell spatial metabolomics in undecalcified bones faces several significant challenges, such as the fragility of bone which often requires decalcification or fixation leading to the degradation or removal of lipids and other molecules and. As such, we describe a method for performing mass spectrometry imaging on undecalcified spine that is compatible with other spatial omics measurements. In brief, we use fresh-freeze rat spines and a system of carboxyl methylcellulose embedding, cryofilm, and polytetrafluoroethylene rollers to maintain tissue integrity, while avoiding signal loss from variations in laser focus and artifacts from traditional tissue processing. This reveals various tissue types and lipidomic profiles of spinal regions at 10 μm spatial resolutions using matrix-assisted laser desorption/ionization mass spectrometry imaging. We expect this method to be adapted and applied to the analysis of spinal cord, shedding light on the mechanistic aspects of cellular heterogeneity, development, and disease pathogenesis underlying different bone-related conditions and diseases. This study furthers the methodology for high spatial metabolomics of spines, as well as adds to the collective efforts to achieve a holistic understanding of diseases via single-cell spatial multi-omics.
Keywords: Lipidomics; imaging; metabolomics; neuroscience; spine.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Little J. P. Chapter 22 - The Spine: Biomechanics and Subject-Specific Finite Element Models. In DHM and Posturography; Scataglini S., Paul G., Eds.; Academic Press, 2019; pp 287–293. 10.1016/B978-0-12-816713-7.00022-2. - DOI
-
- Kayalioglu G. The V, ertebral Column and Spinal Meninges. Spinal Cord 2009, 17–36. 10.1016/B978-0-12-374247-6.50007-9. - DOI
-
- Heise C.; Kayalioglu G. Chapter 13 - Spinal Cord Transmitter Substances. In The Spinal Cord; Watson C., Paxinos G., Kayalioglu G., Eds.; Academic Press: San Diego, 2009; pp 191–208. 10.1016/B978-0-12-374247-6.50017-1. - DOI
-
- Brown A. G. Organization in the Spinal Cord: The Anatomy and Physiology of Identified Neurones; Springer Science & Business Media, 2012.
-
- McDonald J. W.; Belegu V.; Becker D. Chapter 64 - Spinal Cord. In Principles of Tissue Engineering (Fourth Edition); Lanza R., Langer R., Vacanti J., Eds.; Academic Press: Boston, 2014; pp 1353–1373. 10.1016/B978-0-12-398358-9.00064-1. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
