The spread of infectious diseases from a physics perspective
- PMID: 37662617
- PMCID: PMC10469146
- DOI: 10.1093/biomethods/bpad010
The spread of infectious diseases from a physics perspective
Abstract
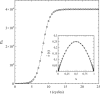
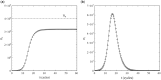
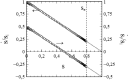
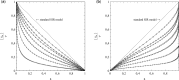
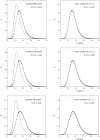
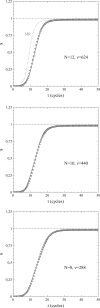
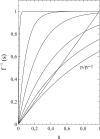
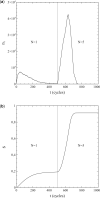
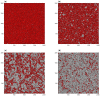
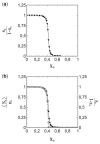
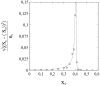
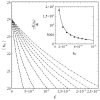
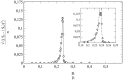
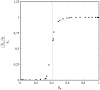
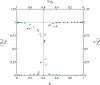
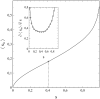
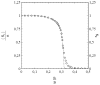
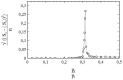
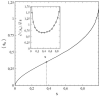
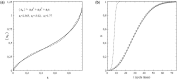
This article deals with the spread of infectious diseases from a physics perspective. It considers a population as a network of nodes representing the population members, linked by network edges representing the (social) contacts of the individual population members. Infections spread along these edges from one node (member) to another. This article presents a novel, modified version of the SIR compartmental model, able to account for typical network effects and percolation phenomena. The model is successfully tested against the results of simulations based on Monte-Carlo methods. Expressions for the (basic) reproduction numbers in terms of the model parameters are presented, and justify some mild criticisms on the widely spread interpretation of reproduction numbers as being the number of secondary infections due to a single active infection. Throughout the article, special emphasis is laid on understanding, and on the interpretation of phenomena in terms of concepts borrowed from condensed-matter and statistical physics, which reveals some interesting analogies. Percolation effects are of particular interest in this respect and they are the subject of a detailed investigation. The concept of herd immunity (its definition and nature) is intensively dealt with as well, also in the context of large-scale vaccination campaigns and waning immunity. This article elucidates how the onset of herd-immunity can be considered as a second-order phase transition in which percolation effects play a crucial role, thus corroborating, in a more pictorial/intuitive way, earlier viewpoints on this matter. An exact criterium for the most relevant form of herd-immunity to occur can be derived in terms of the model parameters. The analyses presented in this article provide insight in how various measures to prevent an epidemic spread of an infection work, how they can be optimized and what potentially deceptive issues have to be considered when such measures are either implemented or scaled down.
Keywords: Covid-19; SIR-Model; mathematical epidemiology; percolation; phase transitions; renormalization.
© The Author(s) 2023. Published by Oxford University Press.
Figures













Similar articles
-
An extended epidemic model with vaccination: Weak-immune SIRVI.Physica A. 2022 Jul 15;598:127429. doi: 10.1016/j.physa.2022.127429. Epub 2022 Apr 22. Physica A. 2022. PMID: 35498560 Free PMC article.
-
Modelling infectious diseases with herd immunity in a randomly mixed population.Sci Rep. 2021 Oct 18;11(1):20574. doi: 10.1038/s41598-021-00013-2. Sci Rep. 2021. PMID: 34663839 Free PMC article.
-
Edge-Based Compartmental Modelling of an SIR Epidemic on a Dual-Layer Static-Dynamic Multiplex Network with Tunable Clustering.Bull Math Biol. 2018 Oct;80(10):2698-2733. doi: 10.1007/s11538-018-0484-5. Epub 2018 Aug 22. Bull Math Biol. 2018. PMID: 30136212 Free PMC article.
-
Epidemics, the Ising-model and percolation theory: A comprehensive review focused on Covid-19.Physica A. 2021 Jul 1;573:125963. doi: 10.1016/j.physa.2021.125963. Epub 2021 Mar 29. Physica A. 2021. PMID: 33814681 Free PMC article. Review.
-
Herd immunity to endemic diseases: Historical concepts and implications for public health policy.J Eval Clin Pract. 2024 Jun;30(4):625-631. doi: 10.1111/jep.13983. Epub 2024 Apr 1. J Eval Clin Pract. 2024. PMID: 38562003 Review.
Cited by
-
Understanding underlying physical mechanism reveals early warning indicators and key elements for adaptive infections disease networks.PNAS Nexus. 2024 Jun 26;3(7):pgae237. doi: 10.1093/pnasnexus/pgae237. eCollection 2024 Jul. PNAS Nexus. 2024. PMID: 39035039 Free PMC article.
References
-
- Kermack WO, McKendrick AG.. Contribution to the mathematical theory of epidemics. Proc R Soc A 1927;115:700–21. - PubMed
-
- Ising E. Beiträge zur theorie des ferromagnetismus. Z Physik 1925;31:253–8.
-
- Stauffer D, Aharony A, Filk T.. Perkolationstheorie (transl.). VCH Verlagsgesellschaft Weinheim, 1995. ISBN 3-527-29334-5.
LinkOut - more resources
Full Text Sources
