Differentially expressed non-coding RNAs and their regulatory networks in liver cancer
- PMID: 37662778
- PMCID: PMC10474437
- DOI: 10.1016/j.heliyon.2023.e19223
Differentially expressed non-coding RNAs and their regulatory networks in liver cancer
Abstract
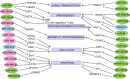
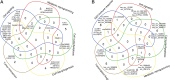
The vast majority of human transcriptome is represented by various types of small RNAs with little or no protein-coding capability referred to as non-coding RNAs (ncRNAs). Functional ncRNAs include microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), which are expressed at very low, but stable and reproducible levels in a variety of cell types. ncRNAs regulate gene expression due to miRNA capability of complementary base pairing with mRNAs, whereas lncRNAs and circRNAs can sponge miRNAs off their target mRNAs to act as competitive endogenous RNAs (ceRNAs). Each miRNA can target multiple mRNAs and a single mRNA can interact with several miRNAs, thereby creating miRNA-mRNA, lncRNA-miRNA-mRNA, and circRNA-miRNA-mRNA regulatory networks. Over the past few years, a variety of differentially expressed miRNAs, lncRNAs, and circRNAs (DEMs, DELs, and DECs, respectively) have been linked to cancer pathogenesis. They can exert both oncogenic and tumor suppressor roles. In this review, we discuss the recent advancements in uncovering the roles of DEMs, DELs, and DECs and their networks in aberrant cell signaling, cell cycle, transcription, angiogenesis, and apoptosis, as well as tumor microenvironment remodeling and metabolic reprogramming during hepatocarcinogenesis. We highlight the potential and challenges in the use of differentially expressed ncRNAs as biomarkers for liver cancer diagnosis and prognosis.
Keywords: Hepatocellular carcinoma; Intrahepatic cholangiocarcinoma; Regulatory networks; circRNA; lncRNA; miRNA.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Arraiano C.M. Regulatory noncoding RNAs: functions and applications in health and disease. FEBS J. 2021;288:6308–6309. 10.1111/febs.16027. Epub 2021. - PubMed
-
- Grillone K., Riillo C., Scionti F., Rocca R., Tradigo G., Guzzi P.H., Alcaro S., Di Martino M.T., Tagliaferri P., Tassone P. Non-coding RNAs in cancer: platforms and strategies for investigating the genomic "dark matter. J. Exp. Clin. Cancer Res. 2020;39:117. doi: 10.1186/s13046-020-01622-x. - DOI - PMC - PubMed
-
- Alexandrov L.B., Kim J., Haradhvala N.J., Huang M.N., Tian Ng A.W., Wu Y., Boot A., Covington K.R., Gordenin D.A., Bergstrom E.N., Islam S.M.A., Lopez-Bigas N., Klimczak L.J., McPherson J.R., Morganella S., Sabarinathan R., Wheeler D.A., Mustonen V., Pcawg Mutational Signatures Working Group, Getz G., Rozen S.G., Stratton M.R., Consortium P.C.A.W.G. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. doi: 10.1038/s41586-020-1943-3. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

