DNA methylation patterns in CD4+ T-cells separate psoriasis patients from healthy controls, and skin psoriasis from psoriatic arthritis
- PMID: 37662940
- PMCID: PMC10472451
- DOI: 10.3389/fimmu.2023.1245876
DNA methylation patterns in CD4+ T-cells separate psoriasis patients from healthy controls, and skin psoriasis from psoriatic arthritis
Abstract
Background: Psoriasis is an autoimmune/inflammatory disorder primarily affecting the skin. Chronic joint inflammation triggers the diagnosis of psoriatic arthritis (PsA) in approximately one-third of psoriasis patients. Although joint disease typically follows the onset of skin psoriasis, in around 15% of cases it is the initial presentation, which can result in diagnostic delays. The pathophysiological mechanisms underlying psoriasis and PsA are not yet fully understood, but there is evidence pointing towards epigenetic dysregulation involving CD4+ and CD8+ T-cells.
Objectives: The aim of this study was to investigate disease-associated DNA methylation patterns in CD4+ T-cells from psoriasis and PsA patients that may represent potential diagnostic and/or prognostic biomarkers.
Methods: PBMCs were collected from 12 patients with chronic plaque psoriasis and 8 PsA patients, and 8 healthy controls. CD4+ T-cells were separated through FACS sorting, and DNA methylation profiling was performed (Illumina EPIC850K arrays). Bioinformatic analyses, including gene ontology (GO) and KEGG pathway analysis, were performed using R. To identify genes under the control of interferon (IFN), the Interferome database was consulted, and DNA Methylation Scores were calculated.
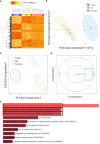
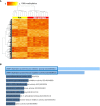
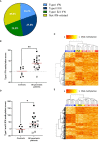
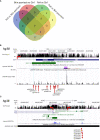
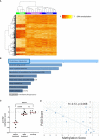
Results: Numbers and proportions of CD4+ T-cell subsets (naïve, central memory, effector memory, CD45RA re-expressing effector memory cells) did not vary between controls, skin psoriasis and PsA patients. 883 differentially methylated positions (DMPs) affecting 548 genes were identified between controls and "all" psoriasis patients. Principal component and partial least-squares discriminant analysis separated controls from skin psoriasis and PsA patients. GO analysis considering promoter DMPs delivered hypermethylation of genes involved in "regulation of wound healing, spreading of epidermal cells", "negative regulation of cell-substrate junction organization" and "negative regulation of focal adhesion assembly". Comparing controls and "all" psoriasis, a majority of DMPs mapped to IFN-related genes (69.2%). Notably, DNA methylation profiles also distinguished skin psoriasis from PsA patients (2,949 DMPs/1,084 genes) through genes affecting "cAMP-dependent protein kinase inhibitor activity" and "cAMP-dependent protein kinase regulator activity". Treatment with cytokine inhibitors (IL-17/TNF) corrected DNA methylation patterns of IL-17/TNF-associated genes, and methylation scores correlated with skin disease activity scores (PASI).
Conclusion: DNA methylation profiles in CD4+ T-cells discriminate between skin psoriasis and PsA. DNA methylation signatures may be applied for quantification of disease activity and patient stratification towards individualized treatment.
Keywords: CD4+ T-cell; biomarker; epigenetics; interferon; methylation; psoriasis; psoriatic arthritis.
Copyright © 2023 Natoli, Charras, Hofmann, Northey, Russ, Schulze, McCann, Abraham and Hedrich.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

