Identification of Zip8-correlated hub genes in pulmonary hypertension by informatic analysis
- PMID: 37663293
- PMCID: PMC10470448
- DOI: 10.7717/peerj.15939
Identification of Zip8-correlated hub genes in pulmonary hypertension by informatic analysis
Abstract
Background: Pulmonary hypertension (PH) is a syndrome characterized by marked remodeling of the pulmonary vasculature and increased pulmonary vascular resistance, ultimately leading to right heart failure and even death. The localization of Zrt/Irt-like Protein 8 (ZIP8, a metal ion transporter, encoded by SLC39A8) was abundantly in microvasculature endothelium and its pivotal role in the lung has been demonstrated. However, the role of Zip8 in PH remains unclear.
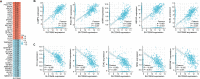
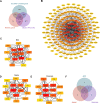
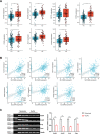
Methods: Bioinformatics analysis was employed to identify SLC39A8 expression patterns and differentially expressed genes (DEGs) between PH patients and normal controls (NC), based on four datasets (GSE24988, GSE113439, GSE117261, and GSE15197) from the Biotechnology Gene Expression Omnibus (NCBI GEO) database. Gene set enrichment analysis (GSEA) was performed to analyze signaling pathways enriched for DEGs. Hub genes were identified by cytoHubba analysis in Cytoscape. Reverse transcriptase-polymerase chain reaction was used to validate SLC39A8 and its correlated metabolic DEGs expression in PH (SU5416/Hypoxia) mice.
Results: SLC39A8 expression was downregulated in PH patients, and this expression pattern was validated in PH (SU5416/Hypoxia) mouse lung tissue. SLC39A8-correlated genes were mainly enriched in the metabolic pathways. Within these SLC39A8-correlated genes, 202 SLC39A8-correlated metabolic genes were screened out, and seven genes were identified as SLC39A8-correlated metabolic hub genes. The expression patterns of hub genes were analyzed between PH patients and controls and further validated in PH mice. Finally, four genes (Fasn, Nsdhl, Acat2, and Acly) were downregulated in PH mice. However, there were no significant differences in the expression of the other three hub genes between PH mice and controls. Of the four genes, Fasn and Acly are key enzymes in fatty acids synthesis, Nsdhl is involved in cholesterol synthesis, and Acat2 is implicated in cholesterol metabolic transformation. Taken together, these results provide novel insight into the role of Zip8 in PH.
Keywords: Differentially expressed genes; Fatty acids; Informatic analysis; Pulmonary hypertension; Zip8.
©2023 Zhao et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


References
-
- Berndt SI, Gustafsson S, Mägi R, Ganna A, Wheeler E, Feitosa MF, Justice AE, Monda KL, Croteau-Chonka DC, Day FR, Esko T, Fall T, Ferreira T, Gentilini D, Jackson AU, Ja Luan, Randall JC, Vedantam S, Willer CJ, Winkler TW, Wood AR, Workalemahu T, Hu Y-J, Lee SH, Liang L, Lin D-Y, Min JL, Neale BM, Thorleifsson G, Yang J, Albrecht E, Amin N, Bragg-Gresham JL, Cadby G, den Heijer M, Eklund N, Fischer K, Goel A, Hottenga J-J, Huffman JE, Jarick I, Johansson Å, Johnson T, Kanoni S, Kleber ME, König IR, Kristiansson K, Kutalik Z, Lamina C, Lecoeur C, Li G, Mangino M, McArdle WL, Medina-Gomez C, Müller-Nurasyid M, Ngwa JS, Nolte IM, Paternoster L, Pechlivanis S, Perola M, Peters MJ, Preuss M, Rose LM, Shi J, Shungin D, Smith AV, Strawbridge RJ, Surakka I, Teumer A, Trip MD, Tyrer J, Van Vliet-Ostaptchouk JV, Vandenput L, Waite LL, Zhao JH, Absher D, Asselbergs FW, Atalay M, Attwood AP, Balmforth AJ, Basart H, Beilby J, Bonnycastle LL, Brambilla P, Bruinenberg M, Campbell H, Chasman DI, Chines PS, Collins FS, Connell JM, Cookson WO, de Faire U, de Vegt F, Dei M, Dimitriou M, Edkins S, Estrada K, Evans DM, Farrall M, Ferrario MM, Ferrières J, Franke L, Frau F, Gejman PV, Grallert H, Grönberg H, Gudnason V, Hall AS, Hall P, Hartikainen A-L, Hayward C, Heard-Costa NL, Heath AC, Hebebrand J, Homuth G, Hu FB, Hunt SE, Hyppönen E, Iribarren C, Jacobs KB, Jansson J-O, Jula A, Kähönen M, Kathiresan S, Kee F, Khaw K-T, Kivimäki M, Koenig W, Kraja AT, Kumari M, Kuulasmaa K, Kuusisto J, Laitinen JH, Lakka TA, Langenberg C, Launer LJ, Lind L, Lindström J, Liu J, Liuzzi A, Lokki M-L, Lorentzon M, Madden PA, Magnusson PK, Manunta P, Marek D, März W, Leach IM, McKnight B, Medland SE, Mihailov E, Milani L, Montgomery GW, Mooser V, Mühleisen TW, Munroe PB, Musk AW, Narisu N, Navis G, Nicholson G, Nohr EA, Ong KK, Oostra BA, Palmer CNA, Palotie A, Peden JF, Pedersen N, Peters A, Polasek O, Pouta A, Pramstaller PP, Prokopenko I, Pütter C, Radhakrishnan A, Raitakari O, Rendon A, Rivadeneira F, Rudan I, Saaristo TE, Sambrook JG, Sanders AR, Sanna S, Saramies J, Schipf S, Schreiber S, Schunkert H, Shin S-Y, Signorini S, Sinisalo J, Skrobek B, Soranzo N, Stančáková A, Stark K, Stephens JC, Stirrups K, Stolk RP, Stumvoll M, Swift AJ, Theodoraki EV, Thorand B. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nature Genetics. 2013;45:501–512. doi: 10.1038/ng.2606. - DOI - PMC - PubMed
-
- Besecker B, Bao S, Bohacova B, Papp A, Sadee W, Knoell DL. The human zinc transporter SLC39A8 (Zip8) is critical in zinc-mediated cytoprotection in lung epithelia. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2008;294:L1127–L1136. doi: 10.1152/ajplung.00057.2008. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

