Sulforaphane prevents the reactivation of HIV-1 by suppressing NFκB signaling
- PMID: 37663574
- PMCID: PMC10469555
- DOI: 10.1016/j.jve.2023.100341
Sulforaphane prevents the reactivation of HIV-1 by suppressing NFκB signaling
Abstract
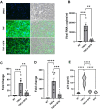
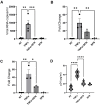
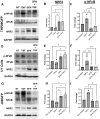
Despite more than 20 years of combination antiretroviral therapy (cART), complete eradication of HIV remains a daunting task. While cART has been very effective in limiting new cycles of infection and keeping viral load below detectable levels with partial restoration of immune functions, it cannot provide a cure. Evidently, the interruption of cART leads to a quick rebound of the viral load within a few weeks. These consistent observations have revealed HIV ability to persist as an undetectable latent reservoir in a variety of tissues that remain insensitive to antiretroviral therapies. The 'Block-and-Lock' approach to drive latent cells into deep latency has emerged as a viable strategy to achieve a functional cure. It entails the development of latency-promoting agents with anti-HIV functions. Recent reports have suggested sulforaphane (SFN), an inducer of NRF-2 (nuclear erythroid 2-related factor 2)-mediated antioxidative signaling, to possess anti-HIV properties by restricting HIV replication at the early stages. However, the effect of SFN on the expression of integrated provirus remains unexplored. We have hypothesized that SFN may promote latency and prevent reactivation. Our results indicate that SFN can render latently infected monocytes and CD4+ T cells resistant to reactivation. SFN treatments antagonized the effects of known latency reactivating agents, tumor necrosis pactor (TNF-α), and phorbol 12-myristate 13-acetate (PMA), and caused a significant reduction in HIV transcription, viral RNA copies, and p24 levels. Furthermore, this block of reactivation was found to be mediated by SFN-induced NRF-2 signaling that specifically decreased the activation of NFκB signaling and thus restricted the HIV-1 promoter (5'LTR) activity. Overall, our study provides compelling evidence to highlight the latency-promoting potential of SFN which could be used in the 'Block-and-Lock' approach to achieve an HIV cure.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

