Characteristics of Autophagy-Related Genes, Diagnostic Models, and Their Correlation with Immune Infiltration in Keratoconus
- PMID: 37663760
- PMCID: PMC10474872
- DOI: 10.2147/JIR.S420164
Characteristics of Autophagy-Related Genes, Diagnostic Models, and Their Correlation with Immune Infiltration in Keratoconus
Abstract
Purpose: Keratoconus (KTCN) is one of the most common degenerative keratopathies, significantly affecting vision and even leading to blindness. This study identifies potential biomarkers of KTCN based on the characterization of autophagy-related genes (ARGs) and the construction of a diagnostic model; and explores their relevance to immune infiltrating cells in KTCN.
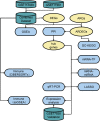
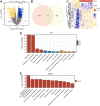
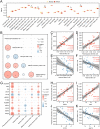
Methods: Gene Expression Omnibus (GEO) data were downloaded and ARGs were acquired from GeneCards and Molecular Signatures Database (MSigDB). Autophagy-related differential expression genes (ARDEGs) were discovered through the integration of differentially expressed genes (DEGs) with ARGs, while hub genes of KTCN were discovered by protein-protein interaction (PPI) network analysis. The probable biological roles of these hub ARDEGs were examined using functional enrichment analysis, and a KTCN diagnostic model was generated using the least absolute shrinkage and selection operator (LASSO) regression analysis. We also employed the CIBERSORTx and ssGSEA algorithms to identify potential regulatory pathways to compare the abundance of immune cell infiltrates and their association with hub genes. Finally, the hub gene expression levels were confirmed using validation datasets as well as blood samples from KTCN and healthy individuals.
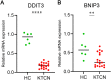
Results: In this study, we identified 12 hub ARDEGs, of which 9 genes were substantially distinct between KTCN patients and normal groups. The LASSO risk score was used to generate the nomogram, and the calibration curve evaluated the model's effective diagnostic performance (C index of 0.961). Patients with KTCN had greater percentages of M2 Macrophages and Gamma delta T cells, according to CIBERSORTx and ssGSEA. The outcomes of the bioinformatics analysis were supported by the DDIT3 and BINP3 expression levels in KTCN patients and healthy controls, according to the qRT-PCR data.
Conclusion: Five biomarkers (CFTR, PLIN2, DDIT3, BAG3, and BNIP3) and diagnostic models offer fresh perspectives on identifying and managing KTCN.
Keywords: autophagy-related genes; bioinformatics analysis; biomarker; diagnostic model; immune infiltration; keratoconus.
© 2023 Liu et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

