The genome sequence of the tree of heaven, Ailanthus altissima (Mill.) Swingle, 1916
- PMID: 37663793
- PMCID: PMC10472066
- DOI: 10.12688/wellcomeopenres.19628.1
The genome sequence of the tree of heaven, Ailanthus altissima (Mill.) Swingle, 1916
Abstract
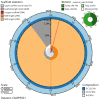
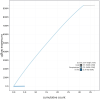
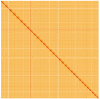
We present a genome assembly from an individual Ailanthus altissima (tree of heaven; Streptophyta; Magnoliopsida; Sapindales; Simaroubaceae). The genome sequence is 939 megabases in span. Most of the assembly is scaffolded into 31 chromosomal pseudomolecules. The mitochondrial and plastid genome assemblies are 661.1 kilobases and 161.1 kilobases long, respectively.
Keywords: Ailanthus altissima; Simaroubaceae; chromosomal; genome sequence; tree of heaven.
Copyright: © 2023 Schley RJ et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


References
-
- Dallas JF, Leitch M, Hulme PE: Microsatellites for tree of heaven ( Ailanthus altissima). Mol Ecol Notes. 2005;5(2):340–342. 10.1111/j.1471-8286.2005.00920.x - DOI
LinkOut - more resources
Full Text Sources

