Demethylation of EHMT1/GLP Protein Reprograms Its Transcriptional Activity and Promotes Prostate Cancer Progression
- PMID: 37663929
- PMCID: PMC10470473
- DOI: 10.1158/2767-9764.CRC-23-0208
Demethylation of EHMT1/GLP Protein Reprograms Its Transcriptional Activity and Promotes Prostate Cancer Progression
Abstract
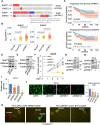
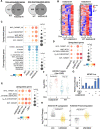
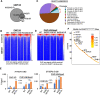
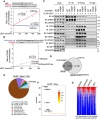
Epigenetic reprogramming, mediated by genomic alterations and dysregulation of histone reader and writer proteins, plays a critical role in driving prostate cancer progression and treatment resistance. However, the specific function and regulation of EHMT1 (also known as GLP) and EHMT2 (also known as G9A), well-known histone 3 lysine 9 methyltransferases, in prostate cancer progression remain poorly understood. Through comprehensive investigations, we discovered that both EHMT1 and EHMT2 proteins have the ability to activate oncogenic transcription programs in prostate cancer cells. Silencing EHMT1/2 or targeting their enzymatic activity with small-molecule inhibitors can markedly decrease prostate cancer cell proliferation and metastasis in vitro and in vivo. In-depth analysis of posttranslational modifications of EHMT1 protein revealed the presence of methylation at lysine 450 and 451 residues in multiple prostate cancer models. Notably, we found that lysine 450 can be demethylated by LSD1. Strikingly, concurrent demethylation of both lysine residues resulted in a rapid and profound expansion of EHMT1's chromatin binding capacity, enabling EHMT1 to reprogram the transcription networks in prostate cancer cells and activate oncogenic signaling pathways. Overall, our studies provide valuable molecular insights into the activity and function of EHMT proteins during prostate cancer progression. Moreover, we propose that the dual-lysine demethylation of EHMT1 acts as a critical molecular switch, triggering the induction of oncogenic transcriptional reprogramming in prostate cancer cells. These findings highlight the potential of targeting EHMT1/2 and their demethylation processes as promising therapeutic strategies for combating prostate cancer progression and overcoming treatment resistance.
Significance: In this study, we demonstrate that EHMT1 and EHMT2 proteins drive prostate cancer development by transcriptionally activating multiple oncogenic pathways. Mechanistically, the chromatin binding of EHMT1 is significantly expanded through demethylation of both lysine 450 and 451 residues, which can serve as a critical molecular switch to induce oncogenic transcriptional reprogramming in prostate cancer cells.
© 2023 The Authors; Published by the American Association for Cancer Research.
Figures




References
-
- Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. . Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367:1187–97. - PubMed
-
- Ge R, Wang Z, Montironi R, Jiang Z, Cheng M, Santoni M, et al. . Epigenetic modulations and lineage plasticity in advanced prostate cancer. Ann Oncol 2020;31:470–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

