Fecal microbial biomarkers combined with multi-target stool DNA test improve diagnostic accuracy for colorectal cancer
- PMID: 37663945
- PMCID: PMC10473925
- DOI: 10.4251/wjgo.v15.i8.1424
Fecal microbial biomarkers combined with multi-target stool DNA test improve diagnostic accuracy for colorectal cancer
Abstract
Background: Colorectal cancer (CRC) is a major global health burden. The current diagnostic tests have shortcomings of being invasive and low accuracy.
Aim: To explore the combination of intestinal microbiome composition and multi-target stool DNA (MT-sDNA) test in the diagnosis of CRC.
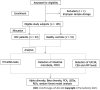
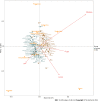
Methods: We assessed the performance of the MT-sDNA test based on a hospital clinical trial. The intestinal microbiota was tested using 16S rRNA gene sequencing. This case-control study enrolled 54 CRC patients and 51 healthy controls. We identified biomarkers of bacterial structure, analyzed the relationship between different tumor markers and the relative abundance of related flora components, and distinguished CRC patients from healthy subjects by the linear discriminant analysis effect size, redundancy analysis, and random forest analysis.
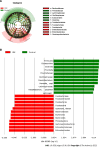
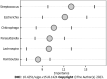
Results: MT-sDNA was associated with Bacteroides. MT-sDNA and carcinoembryonic antigen (CEA) were positively correlated with the existence of Parabacteroides, and alpha-fetoprotein (AFP) was positively associated with Faecalibacterium and Megamonas. In the random forest model, the existence of Streptococcus, Escherichia, Chitinophaga, Parasutterella, Lachnospira, and Romboutsia can distinguish CRC from health controls. The diagnostic accuracy of MT-sDNA combined with the six genera and CEA in the diagnosis of CRC was 97.1%, with a sensitivity and specificity of 98.1% and 92.3%, respectively.
Conclusion: There is a positive correlation of MT-sDNA, CEA, and AFP with intestinal microbiome. Eight biomarkers including six genera of gut microbiota, MT-sDNA, and CEA showed a prominent sensitivity and specificity for CRC prediction, which could be used as a non-invasive method for improving the diagnostic accuracy for this malignancy.
Keywords: Colorectal cancer; Diagnostic model; Gut microbiome; Multi-target stool DNA test; Tumor biomarker.
©The Author(s) 2023. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: All the authors report no relevant conflicts of interest for this article.
Figures

References
-
- Wu W, Huang J, Yang Y, Gu K, Luu HN, Tan S, Yang C, Fu J, Bao P, Ying T, Withers M, Mao D, Chen S, Gong Y, Wong MCS, Xu W. Adherence to colonoscopy in cascade screening of colorectal cancer: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2022;37:620–631. - PubMed
LinkOut - more resources
Full Text Sources

