Transcriptomic and proteomic assessment of tocilizumab response in a randomized controlled trial of patients hospitalized with COVID-19
- PMID: 37664617
- PMCID: PMC10470387
- DOI: 10.1016/j.isci.2023.107597
Transcriptomic and proteomic assessment of tocilizumab response in a randomized controlled trial of patients hospitalized with COVID-19
Abstract
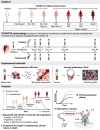
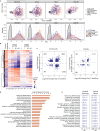
High interleukin (IL)-6 levels are associated with greater COVID-19 severity. IL-6 receptor blockade by tocilizumab (anti-IL6R; Actemra) is used globally for the treatment of severe COVID-19, yet a molecular understanding of the therapeutic benefit remains unclear. We characterized the immune profile and identified cellular and molecular pathways modified by tocilizumab in peripheral blood samples from patients enrolled in the COVACTA study, a phase 3, randomized, double-blind, placebo-controlled trial of the efficacy and safety of tocilizumab in hospitalized patients with severe COVID-19. We identified markers of inflammation, lymphopenia, myeloid dysregulation, and organ injury that predict disease severity and clinical outcomes. Proteomic analysis confirmed a pharmacodynamic effect for tocilizumab and identified novel pharmacodynamic biomarkers. Transcriptomic analysis revealed that tocilizumab treatment leads to faster resolution of lymphopenia and myeloid dysregulation associated with severe COVID-19, indicating greater anti-inflammatory activity relative to placebo and potentially leading to faster recovery in patients hospitalized with COVID-19.
Keywords: Omics; Pharmaceutical science; Virology.
© 2023 The Authors.
Conflict of interest statement
H.S., J.A.H., C.M.R., A.Q., O.O., J.M., F.C., M.B., L.T., A.R., and R.N.B. are employees of Roche/Genentech and hold stock and/or stock options in Roche/Genentech. A.T. is an employee of Roche/Genentech. F.C. has a patent pending to Genentech for biomarkers for predicting a response to an interleukin (IL)-6 antagonist (P36367-US). L.T. is an author of a patent Method for Treating Pneumonia, including COVID-19 Pneumonia with an IL-6 Antagonist pending, owned by Genentech/Roche. A.R. is a co-founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas Therapeutics and, until July 31, 2020, was a scientific advisory board member of ThermoFisher Scientific, Syros Pharmaceuticals, Asimov, and Neogene Therapeutics. A.R. is a named inventor on multiple patents related to single cell and spatial genomics filed by or issued to the Broad Institute. I.O.R. has nothing to declare.
Figures





Similar articles
-
Treatment of severely ill COVID-19 patients with anti-interleukin drugs (COV-AID): A structured summary of a study protocol for a randomised controlled trial.Trials. 2020 Jun 3;21(1):468. doi: 10.1186/s13063-020-04453-5. Trials. 2020. PMID: 32493441 Free PMC article.
-
Safety and Efficacy of Imatinib for Hospitalized Adults with COVID-19: A structured summary of a study protocol for a randomised controlled trial.Trials. 2020 Oct 28;21(1):897. doi: 10.1186/s13063-020-04819-9. Trials. 2020. PMID: 33115543 Free PMC article.
-
A prospective, randomised, double blind placebo-controlled trial to evaluate the efficacy and safety of tocilizumab in patients with severe COVID-19 pneumonia (TOC-COVID): A structured summary of a study protocol for a randomised controlled trial.Trials. 2020 Jun 3;21(1):470. doi: 10.1186/s13063-020-04447-3. Trials. 2020. PMID: 32493514 Free PMC article.
-
Tocilizumab in SARS-CoV-2 Patients with the Syndrome of Cytokine Storm: A Narrative Review.Rev Recent Clin Trials. 2021;16(2):138-145. doi: 10.2174/1574887115666200917110954. Rev Recent Clin Trials. 2021. PMID: 32940187 Review.
-
Efficacy and safety of tocilizumab in COVID-19 patients: a living systematic review and meta-analysis, first update.Clin Microbiol Infect. 2021 Aug;27(8):1076-1082. doi: 10.1016/j.cmi.2021.04.019. Epub 2021 Apr 27. Clin Microbiol Infect. 2021. PMID: 33915284 Free PMC article.
Cited by
-
Systemic cytokines drive conserved severity-associated myeloid responses across bacterial and viral infections.Commun Biol. 2025 Jul 23;8(1):1096. doi: 10.1038/s42003-025-08407-y. Commun Biol. 2025. PMID: 40702253 Free PMC article.
-
A myeloid program associated with COVID-19 severity is decreased by therapeutic blockade of IL-6 signaling.iScience. 2023 Sep 1;26(10):107813. doi: 10.1016/j.isci.2023.107813. eCollection 2023 Oct 20. iScience. 2023. PMID: 37810211 Free PMC article.
-
An exploratory analysis of associations of genetic variation with the efficacy of tocilizumab in severe COVID-19 patients. A pharmacogenetic study based on next-generation sequencing.Front Pharmacol. 2024 Sep 13;15:1426826. doi: 10.3389/fphar.2024.1426826. eCollection 2024. Front Pharmacol. 2024. PMID: 39346556 Free PMC article.
-
The Functional Roles of MDSCs in Severe COVID-19 Pathogenesis.Viruses. 2023 Dec 23;16(1):27. doi: 10.3390/v16010027. Viruses. 2023. PMID: 38257728 Free PMC article. Review.
-
Immunobiological effects of tocilizumab across respiratory subphenotypes in COVID-19 ARDS.Intensive Care Med Exp. 2025 Jul 9;13(1):70. doi: 10.1186/s40635-025-00779-z. Intensive Care Med Exp. 2025. PMID: 40632330 Free PMC article.
References
-
- WHO Rapid Evidence Appraisal for COVID-19 Therapies REACT Working Group. Shankar-Hari M., Vale C.L., Godolphin P.J., Fisher D., Higgins J.P.T., Spiga F., Savovic J., Tierney J., Baron G., et al. Association between administration of IL-6 antagonists and mortality among patients hospitalized for COVID-19: A meta-analysis. JAMA. 2021;326:499–518. doi: 10.1001/jama.2021.11330. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

