EpiGe: A machine-learning strategy for rapid classification of medulloblastoma using PCR-based methyl-genotyping
- PMID: 37664618
- PMCID: PMC10470382
- DOI: 10.1016/j.isci.2023.107598
EpiGe: A machine-learning strategy for rapid classification of medulloblastoma using PCR-based methyl-genotyping
Abstract
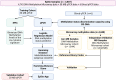
Molecular classification of medulloblastoma is critical for the treatment of this brain tumor. Array-based DNA methylation profiling has emerged as a powerful approach for brain tumor classification. However, this technology is currently not widely available. We present a machine-learning decision support system (DSS) that enables the classification of the principal molecular groups-WNT, SHH, and non-WNT/non-SHH-directly from quantitative PCR (qPCR) data. We propose a framework where the developed DSS appears as a user-friendly web-application-EpiGe-App-that enables automated interpretation of qPCR methylation data and subsequent molecular group prediction. The basis of our classification strategy is a previously validated six-cytosine signature with subgroup-specific methylation profiles. This reduced set of markers enabled us to develop a methyl-genotyping assay capable of determining the methylation status of cytosines using qPCR instruments. This study provides a comprehensive approach for rapid classification of clinically relevant medulloblastoma groups, using readily accessible equipment and an easy-to-use web-application.t.
Keywords: Cancer; Health technology; Machine learning.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Capper D., Stichel D., Sahm F., Jones D.T.W., Schrimpf D., Sill M., Schmid S., Hovestadt V., Reuss D.E., Koelsche C., et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 2018;136:181–210. doi: 10.1007/s00401-018-1879-y. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

