Development of a loop-mediated isothermal amplification (LAMP)-based electrochemical test for rapid detection of SARS-CoV-2
- PMID: 37664622
- PMCID: PMC10470312
- DOI: 10.1016/j.isci.2023.107570
Development of a loop-mediated isothermal amplification (LAMP)-based electrochemical test for rapid detection of SARS-CoV-2
Abstract
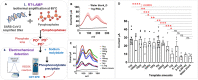
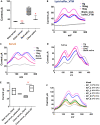
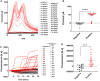
Rapid, reliable, sensitive, portable, and accurate diagnostics are required to control disease outbreaks such as COVID-19 that pose an immense burden on human health and the global economy. Here we developed a loop-mediated isothermal amplification (LAMP)-based electrochemical test for the detection of SARS-CoV-2 that causes COVID-19. The test is based on the oxidation-reduction reaction between pyrophosphates (generated from positive LAMP reaction) and molybdate that is detected by cyclic voltammetry using inexpensive and disposable carbon screen printed electrodes. Our test showed higher sensitivity (detecting as low as 5.29 RNA copies/μL) compared to the conventional fluorescent reverse transcriptase (RT)-LAMP. We validated our tests using human serum and saliva spiked with SARS-CoV-2 RNA and clinical (saliva and nasal-pharyngeal) swab samples demonstrating 100% specificity and 93.33% sensitivity. Our assay provides a rapid, specific, and sensitive test with an electrochemical readout in less than 45 min that could be adapted for point-of-care settings.
Keywords: Devices; Diagnostic technique in health technology.
© 2023 The Authors.
Conflict of interest statement
All affiliations are listed on the title page of the manuscript. All funding sources for this study are listed in the “acknowledgments” section of the manuscript. We, the authors and our immediate family members, have no financial interests to declare. We, the authors, and our immediate family members, have no positions to declare and are not members of the journal’s advisory board. We declare that we have a patent application in process related to this work.
Figures


References
-
- World Health Organization . 2022. Weekly Operational Update on COVID-19. Issue No. 91.https://www.who.int/publications/m/item/weekly-operational-update-on-cov...
-
- Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis, and therapies: Structural genomics approach. Biochim. Biophys. Acta, Mol. Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

