Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA
- PMID: 37665001
- PMCID: PMC10570014
- DOI: 10.1093/nar/gkad724
Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA
Abstract
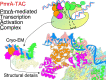
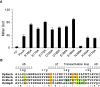
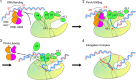
PmrA, an OmpR/PhoB-family response regulator, triggers gene transcription responsible for polymyxin resistance in bacteria by recognizing promoters where the canonical-35 element is replaced by the pmra-box, representing the PmrA recognition sequence. Here, we report a cryo-electron microscopy (cryo-EM) structure of a bacterial PmrA-dependent transcription activation complex (TAC) containing a PmrA dimer, an RNA polymerase σ70 holoenzyme (RNAPH) and the pbgP promoter DNA. Our structure reveals that the RNAPH mainly contacts the PmrA C-terminal DNA-binding domain (DBD) via electrostatic interactions and reorients the DBD three base pairs upstream of the pmra-box, resulting in a dynamic TAC conformation. In vivo assays show that the substitution of the DNA-recognition residue eliminated its transcriptional activity, while variants with altered RNAPH-interacting residues resulted in enhanced transcriptional activity. Our findings suggest that both PmrA recognition-induced DNA distortion and PmrA promoter escape play crucial roles in its transcriptional activation.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Gunn J.S. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 2008; 16:284–290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

