Ets-1 transcription factor regulates glial cell regeneration and function in planarians
- PMID: 37665145
- PMCID: PMC10508700
- DOI: 10.1242/dev.201666
Ets-1 transcription factor regulates glial cell regeneration and function in planarians
Abstract
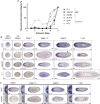
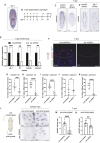
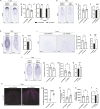
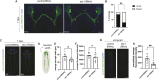
Glia play multifaceted roles in nervous systems in response to injury. Depending on the species, extent of injury and glial cell type in question, glia can help or hinder the regeneration of neurons. Studying glia in the context of successful regeneration could reveal features of pro-regenerative glia that could be exploited for new human therapies. Planarian flatworms completely regenerate their nervous systems after injury - including glia - and thus provide a strong model system for exploring glia in the context of regeneration. Here, we report that planarian glia regenerate after neurons, and that neurons are required for correct glial numbers and localization during regeneration. We also identify the planarian transcription factor-encoding gene ets-1 as a key regulator of glial cell maintenance and regeneration. Using ets-1 (RNAi) to perturb glia, we show that glial loss is associated with altered neuronal gene expression, impeded animal movement and impaired nervous system architecture - particularly within the neuropil. Importantly, our work reveals the inter-relationships of glia and neurons in the context of robust neural regeneration.
Keywords: S. mediterranea; ETS; Glia; Nervous system; Planarian; Regeneration.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



Similar articles
-
Coordinated neuron-glia regeneration through Notch signaling in planarians.PLoS Genet. 2025 Jan 27;21(1):e1011577. doi: 10.1371/journal.pgen.1011577. eCollection 2025 Jan. PLoS Genet. 2025. PMID: 39869602 Free PMC article.
-
Hedgehog signaling regulates gene expression in planarian glia.Elife. 2016 Sep 9;5:e16996. doi: 10.7554/eLife.16996. Elife. 2016. PMID: 27612382 Free PMC article.
-
Djsnon, a downstream gene of Djfoxk1, is required for the regeneration of the planarian central nervous system.Biochem Biophys Res Commun. 2023 Feb 5;643:8-15. doi: 10.1016/j.bbrc.2022.12.074. Epub 2022 Dec 24. Biochem Biophys Res Commun. 2023. PMID: 36584589
-
[Molecular mechanism of brain regeneration and reconstruction of dopaminergic neural network in planarians].Brain Nerve. 2008 Apr;60(4):307-17. Brain Nerve. 2008. PMID: 18421972 Review. Japanese.
-
[Regeneration of planarians: experimental object].Ontogenez. 2015 Jan-Feb;46(1):3-12. Ontogenez. 2015. PMID: 25898529 Review. Russian.
Cited by
-
Coordinated neuron-glia regeneration through Notch signaling in planarians.PLoS Genet. 2025 Jan 27;21(1):e1011577. doi: 10.1371/journal.pgen.1011577. eCollection 2025 Jan. PLoS Genet. 2025. PMID: 39869602 Free PMC article.
-
Combinatorial mechanisms specify cellular location and neurotransmitter identity during regeneration of planarian neurons.bioRxiv [Preprint]. 2025 May 25:2025.05.23.655781. doi: 10.1101/2025.05.23.655781. bioRxiv. 2025. PMID: 40661405 Free PMC article. Preprint.
-
A parenchymal niche regulates pluripotent stem cell function in planarians.bioRxiv [Preprint]. 2025 Aug 2:2025.08.01.668211. doi: 10.1101/2025.08.01.668211. bioRxiv. 2025. PMID: 40766428 Free PMC article. Preprint.
-
Utility of the continuous spectrum formed by pathological states in characterizing disease properties.NPJ Syst Biol Appl. 2025 Aug 29;11(1):100. doi: 10.1038/s41540-025-00579-x. NPJ Syst Biol Appl. 2025. PMID: 40883323 Free PMC article.
-
3D reconstruction of neuronal allometry and neuromuscular projections in asexual planarians using expansion tiling light sheet microscopy.Elife. 2025 Mar 28;13:RP101103. doi: 10.7554/eLife.101103. Elife. 2025. PMID: 40152910 Free PMC article.
References
-
- Alesci, A., Pergolizzi, S., Lo Cascio, P., Fumia, A. and Lauriano, E. R. (2022). Neuronal regeneration: vertebrates comparative overview and new perspectives for neurodegenerative diseases. Acta Zool. 103, 129-140. 10.1111/azo.12397 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

