Listeria monocytogenes infection in pregnant macaques alters the maternal gut microbiome†
- PMID: 37665249
- PMCID: PMC10651077
- DOI: 10.1093/biolre/ioad104
Listeria monocytogenes infection in pregnant macaques alters the maternal gut microbiome†
Abstract
Objectives: The bacterium Listeria monocytogenes (Lm) is associated with adverse pregnancy outcomes. Infection occurs through consumption of contaminated food that is disseminated to the maternal-fetal interface. The influence on the gastrointestinal microbiome during Lm infection remains unexplored in pregnancy. The objective of this study was to determine the impact of listeriosis on the gut microbiota of pregnant macaques.
Methods: A non-human primate model of listeriosis in pregnancy has been previously described. Both pregnant and non-pregnant cynomolgus macaques were inoculated with Lm and bacteremia and fecal shedding were monitored for 14 days. Non-pregnant animal tissues were collected at necropsy to determine bacterial burden, and fecal samples from both pregnant and non-pregnant animals were evaluated by 16S rRNA next-generation sequencing.
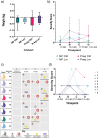
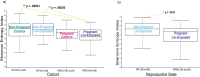
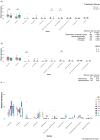
Results: Unlike pregnant macaques, non-pregnant macaques did not exhibit bacteremia, fecal shedding, or tissue colonization by Lm. Dispersion of Lm during pregnancy was associated with a significant decrease in alpha diversity of the host gut microbiome, compared to non-pregnant counterparts. The combined effects of pregnancy and listeriosis were associated with a significant loss in microbial richness, although there were increases in some genera and decreases in others.
Conclusions: Although pregnancy alone is not associated with gut microbiome disruption, we observed dysbiosis with listeriosis during pregnancy. The macaque model may provide an understanding of the roles that pregnancy and the gut microbiota play in the ability of Lm to establish intestinal infection and disseminate throughout the host, thereby contributing to adverse pregnancy outcomes and risk to the developing fetus.
Keywords: bacterial infection; listeriosis; microbiome; non-pregnant; pregnancy; primate.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for the Study of Reproduction. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures





References
-
- Pouillot R, Hoelzer K, Jackson KA, Henao OL, Silk BJ. Relative risk of listeriosis in Foodborne Diseases Active Surveillance Network (FoodNet) sites according to age, pregnancy, and ethnicity. Clin Infect Dis 2012; 54:S405–S410. - PubMed
-
- Thønnings S, Knudsen JD, Schønheyder HC, Søgaard M, Arpi M, Gradel KO, Østergaard C, Østergaard C, Arpi M, Gradel KO, Jensen US, Thønnings S, et al. Antibiotic treatment and mortality in patients with Listeria monocytogenes meningitis or bacteraemia. Clin Microbiol Infect 2016; 22:725–730. - PubMed
-
- Olaimat AN, Al-Holy MA, Shahbaz HM, Al-Nabulsi AA, Abu Ghoush MH, Osaili TM, Ayyash MM, Holley RA. Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: a comprehensive review. Compr Rev Food Sci Food Saf 2018; 17:1277–1292. - PubMed
-
- Sosnowski M, Lachtara B, Wieczorek K, Osek J. Antimicrobial resistance and genotypic characteristics of listeria monocytogenes isolated from food in Poland. Int J Food Microbiol 2019; 289:1–6. - PubMed
-
- Baquero F, Lanza VF, Duval M, Coque TM. Ecogenetics of antibiotic resistance in Listeria monocytogenes. Mol Microbiol 2020; 113:570–579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

