Identification and validation of a prognostic signature of cuproptosis-related genes for esophageal squamous cell carcinoma
- PMID: 37665670
- PMCID: PMC10522377
- DOI: 10.18632/aging.205012
Identification and validation of a prognostic signature of cuproptosis-related genes for esophageal squamous cell carcinoma
Abstract
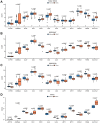
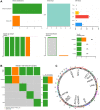
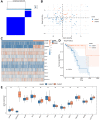
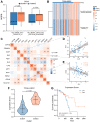
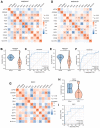
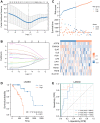
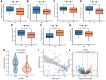
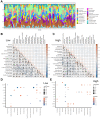
Esophageal squamous cell carcinoma (ESCC) is a highly lethal form of cancer. Cuproptosis is a recently discovered form of regulated cell death. However, its significance in ESCC remains largely unknown. In this study, we observed significant expression differences in most of the 12 cuproptosis-related genes (CRGs) in the TCGA-ESCC dataset, which was validated using GSE20347, GSE38129, and individual ESCC datasets. We were able to divide patients in the TCGA-ESCC cohort into two subgroups based on disease, and found significant differences in survivor outcomes and biological functions between these subgroups. Additionally, we identified 11 prognosis-related genes from the 12 CRGs using LASSO COX regression analysis and constructed a CRGs signature for ESCC. Patients were categorized into high- and low-risk subgroups based on their median risk score, with those in the high-risk subgroup having significantly worse overall survival than those in the low-risk subgroup. The CRGs signature was also highly accurate in predicting prognosis and survival outcomes. Univariate and multivariate Cox regression analyses revealed that 8 of the 11 CRGs were independent prognostic factors for predicting survival in ESCC patients. Furthermore, our nomogram performed well and could serve as a useful tool for predicting prognosis. Finally, our risk model was found to be relevant to the sensitivity of targeted agents and immune infiltration. Functional enrichment analysis demonstrated that the risk model was associated with biological pathways of tumor migration and invasion. In summary, our study may provide a promising prognostic signature based on CRGs and offers potential targets for personalized therapy.
Keywords: cuproptosis; esophageal squamous cell carcinoma; nomogram; prognostic signature; risk score.
Conflict of interest statement
Figures


Similar articles
-
Identification of cuproptosis-related subtypes, construction of a prognosis model, and tumor microenvironment landscape in gastric cancer.Front Immunol. 2022 Nov 21;13:1056932. doi: 10.3389/fimmu.2022.1056932. eCollection 2022. Front Immunol. 2022. PMID: 36479114 Free PMC article.
-
Prognostic value and immune landscapes of cuproptosis-related lncRNAs in esophageal squamous cell carcinoma.Aging (Albany NY). 2023 Oct 6;15(19):10473-10500. doi: 10.18632/aging.205089. Epub 2023 Oct 6. Aging (Albany NY). 2023. PMID: 37812189 Free PMC article.
-
A novel cuproptosis-related prognostic gene signature and validation of differential expression in hepatocellular carcinoma.Front Pharmacol. 2023 Jan 10;13:1081952. doi: 10.3389/fphar.2022.1081952. eCollection 2022. Front Pharmacol. 2023. PMID: 36703728 Free PMC article.
-
Identification of a novel cuproptosis-related gene signature and integrative analyses in patients with lower-grade gliomas.Front Immunol. 2022 Aug 15;13:933973. doi: 10.3389/fimmu.2022.933973. eCollection 2022. Front Immunol. 2022. PMID: 36045691 Free PMC article. Review.
-
Cuproptosis in lung cancer: therapeutic options and prognostic models.Apoptosis. 2024 Oct;29(9-10):1393-1398. doi: 10.1007/s10495-024-01978-x. Epub 2024 May 12. Apoptosis. 2024. PMID: 38735011 Review.
References
-
- Shen L, Kato K, Kim SB, Ajani JA, Zhao K, He Z, Yu X, Shu Y, Luo Q, Wang J, Chen Z, Niu Z, Zhang L, et al., and RATIONALE-302 Investigators. Tislelizumab Versus Chemotherapy as Second-Line Treatment for Advanced or Metastatic Esophageal Squamous Cell Carcinoma (RATIONALE-302): A Randomized Phase III Study. J Clin Oncol. 2022; 40:3065–76. 10.1200/JCO.21.01926 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

