Genome-resolved correlation mapping links microbial community structure to metabolic interactions driving methane production from wastewater
- PMID: 37666802
- PMCID: PMC10477309
- DOI: 10.1038/s41467-023-40907-5
Genome-resolved correlation mapping links microbial community structure to metabolic interactions driving methane production from wastewater
Abstract
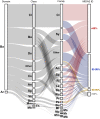
Anaerobic digestion of municipal mixed sludge produces methane that can be converted into renewable natural gas. To improve economics of this microbial mediated process, metabolic interactions catalyzing biomass conversion to energy need to be identified. Here, we present a two-year time series associating microbial metabolism and physicochemistry in a full-scale wastewater treatment plant. By creating a co-occurrence network with thousands of time-resolved microbial populations from over 100 samples spanning four operating configurations, known and novel microbial consortia with potential to drive methane production were identified. Interactions between these populations were further resolved in relation to specific process configurations by mapping metagenome assembled genomes and cognate gene expression data onto the network. Prominent interactions included transcriptionally active Methanolinea methanogens and syntrophic benzoate oxidizing Syntrophorhabdus, as well as a Methanoregulaceae population and putative syntrophic acetate oxidizing bacteria affiliated with Bateroidetes (Tenuifilaceae) expressing the glycine cleavage bypass of the Wood-Ljungdahl pathway.
© 2023. Springer Nature Limited.
Conflict of interest statement
S.J.H. is a co-founder of Koonkie Inc., a bioinformatics consulting company that designs and provides scalable algorithmic and data analytics solutions in the cloud. The remaining authors declare no competing interests.
Figures



References
-
- Adekunle KF, Okolie JA. A review of biochemical process of anaerobic digestion. Adv. Biosci. Biotechnol. 2015;06:205.
-
- Jimenez J, et al. Instrumentation and control of anaerobic digestion processes: a review and some research challenges. Rev. Environ. Sci. Biotechnol. 2015;14:615–648.

