Symmetric inheritance of parental histones contributes to safeguarding the fate of mouse embryonic stem cells during differentiation
- PMID: 37666989
- PMCID: PMC10777717
- DOI: 10.1038/s41588-023-01477-w
Symmetric inheritance of parental histones contributes to safeguarding the fate of mouse embryonic stem cells during differentiation
Abstract
Parental histones, the carriers of posttranslational modifications, are deposited evenly onto the replicating DNA of sister chromatids in a process dependent on the Mcm2 subunit of DNA helicase and the Pole3 subunit of leading-strand DNA polymerase. The biological significance of parental histone propagation remains unclear. Here we show that Mcm2-mutated or Pole3-deleted mouse embryonic stem cells (ESCs) display aberrant histone landscapes and impaired neural differentiation. Mutation of the Mcm2 histone-binding domain causes defects in pre-implantation development and embryonic lethality. ESCs with biased parental histone transfer exhibit increased epigenetic heterogeneity, showing altered histone variant H3.3 and H3K27me3 patterning at genomic sites regulating differentiation genes. Our results indicate that the lagging strand pattern of H3.3 leads to the redistribution of H3K27me3 in Mcm2-2A ESCs. We demonstrate that symmetric parental histone deposition to sister chromatids contributes to cellular differentiation and development.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

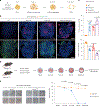



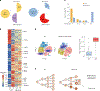

Comment in
-
Asymmetric inheritance of parental histones leads to differentiation defects in mouse embryonic stem cells.Innov Life. 2024 Mar 8;2(1):100047. doi: 10.59717/j.xinn-life.2024.100047. Epub 2024 Jan 9. Innov Life. 2024. PMID: 38601056 Free PMC article. No abstract available.
References
-
- Stillman B Histone modifications: insights into their influence on gene expression. Cell 175, 6–9 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

