Assessing in vivo the impact of gene context on transcription through DNA supercoiling
- PMID: 37667073
- PMCID: PMC10570042
- DOI: 10.1093/nar/gkad688
Assessing in vivo the impact of gene context on transcription through DNA supercoiling
Abstract
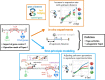
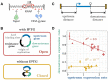
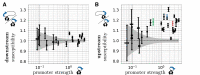
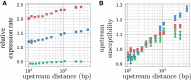
Gene context can have significant impact on gene expression but is currently not integrated in quantitative models of gene regulation despite known biophysical principles and quantitative in vitro measurements. Conceptually, the simplest gene context consists of a single gene framed by two topological barriers, known as the twin transcriptional-loop model, which illustrates the interplay between transcription and DNA supercoiling. In vivo, DNA supercoiling is additionally modulated by topoisomerases, whose modus operandi remains to be quantified. Here, we bridge the gap between theory and in vivo properties by realizing in Escherichia coli the twin transcriptional-loop model and by measuring how gene expression varies with promoters and distances to the topological barriers. We find that gene expression depends on the distance to the upstream barrier but not to the downstream barrier, with a promoter-dependent intensity. We rationalize these findings with a first-principle biophysical model of DNA transcription. Our results are explained if TopoI and gyrase both act specifically, respectively upstream and downstream of the gene, with antagonistic effects of TopoI, which can repress initiation while facilitating elongation. Altogether, our work sets the foundations for a systematic and quantitative description of the impact of gene context on gene regulation.
Plain language summary
The context of genes, particularly the arrangement of neighboring genes along the DNA, exerts an important impact on their expression. However, predicting this impact remains challenging due to the complex interplay of concurrent mechanisms. To gain a quantitative understanding, we experimentally implemented the simplest possible theoretical model, isolating a gene from its neighboring genes. This allowed us to investigate the role of DNA’s mechanical and topological properties, along with the enzymes that shape these properties, including RNA polymerases and topoisomerases. Comparison of the experimental results to a mathematical model based on physical principles allowed us to parametrize the operating mode of topoisomerases. Our work paves the way towards a systematic understanding of the role of gene context in gene expression.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

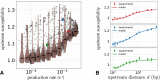
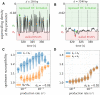
References
-
- Browning D.F., Busby S.J.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. - PubMed
-
- Miravet-Verde S., Lloréns-Rico V., Serrano L.. Alternative transcriptional regulation in genome-reduced bacteria. Curr. Opin. Microbiol. 2017; 39:89–95. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

