Spatial multimodal analysis of transcriptomes and metabolomes in tissues
- PMID: 37667091
- PMCID: PMC11251988
- DOI: 10.1038/s41587-023-01937-y
Spatial multimodal analysis of transcriptomes and metabolomes in tissues
Abstract
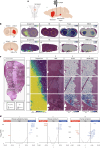
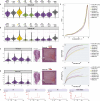
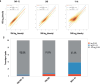
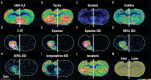
We present a spatial omics approach that combines histology, mass spectrometry imaging and spatial transcriptomics to facilitate precise measurements of mRNA transcripts and low-molecular-weight metabolites across tissue regions. The workflow is compatible with commercially available Visium glass slides. We demonstrate the potential of our method using mouse and human brain samples in the context of dopamine and Parkinson's disease.
© 2023. The Author(s).
Conflict of interest statement
M.V., R.M., L.L., M.N. and J.L. are scientific consultants for 10X Genomics. The other authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
- KAW 2018.172/Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)
- 2022-03984/Vetenskapsrådet (Swedish Research Council)
- 2021-03293/Vetenskapsrådet (Swedish Research Council)
- 2022-04198/Vetenskapsrådet (Swedish Research Council)
- 2020-06182/Vetenskapsrådet (Swedish Research Council)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

