I am better than I look: genome based safety assessment of the probiotic Lactiplantibacillus plantarum IS-10506
- PMID: 37667166
- PMCID: PMC10478331
- DOI: 10.1186/s12864-023-09495-y
I am better than I look: genome based safety assessment of the probiotic Lactiplantibacillus plantarum IS-10506
Abstract
Background: Safety of probiotic strains that are used in human and animal trials is a prerequisite. Genome based safety assessment of probiotics has gained popularity due its cost efficiency and speed, and even became a part of national regulation on foods containing probiotics in Indonesia. However, reliability of the safety assessment based only on a full genome sequence is not clear. Here, for the first time, we sequenced, assembled, and analysed the genome of the probiotic strain Lactiplantibacillus plantarum IS-10506, that was isolated from dadih, a traditional fermented buffalo milk. The strain has already been used as a probiotic for more than a decade, and in several clinical trials proven to be completely safe.
Methods: The genome of the probiotic strain L. plantarum IS-10506 was sequenced using Nanopore sequencing technology, assembled, annotated and screened for potential harmful (PH) and beneficial genomic features. The presence of the PH features was assessed from general annotation, as well as with the use of specialised tools. In addition, PH regions in the genome were compared to all other probiotic and non-probiotic L. plantarum strains available in the NCBI RefSeq database.
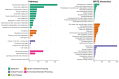
Results: For the first time, a high-quality complete genome of L. plantarum IS-10506 was obtained, and an extensive search for PH and a beneficial signature was performed. We discovered a number of PH features within the genome of L. plantarum IS-10506 based on the general annotation, including various antibiotic resistant genes (AMR); however, with a few exceptions, bioinformatics tools specifically developed for AMR detection did not confirm their presence. We further demonstrated the presence of the detected PH genes across multiple L. plantarum strains, including probiotics, and overall high genetic similarities between strains.
Conclusion: The genome of L. plantarum IS-10506 is predicted to have several PH features. However, the strain has been utilized as a probiotic for over a decade in several clinical trials without any adverse effects, even in immunocompromised children with HIV infection and undernourished children. This implies the presence of PH feature signatures within the probiotic genome does not necessarily indicate their manifestation during administration. Importantly, specialized tools for the search of PH features were found more robust and should be preferred over manual searches in a general annotation.
Keywords: Comparative genomics; Genome; Lactiplantibacillus plantarum; Safety assessment.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- FAO: Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. : Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. In.; 2001.
-
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–14. doi: 10.1038/nrgastro.2014.66. - DOI - PubMed
-
- Akuzawa R, Miura T, Surono I. Asian fermented milks. Encyclopedia of Dairy Science. 2011;2:507–11. doi: 10.1016/B978-0-12-374407-4.00186-2. - DOI
-
- Akuzawa R, Surono IS. Fermented milks of Asia. In: Encyclopedia of dairy sciences Edited by Roginski H, Fuquay JW, Fox PF. London, UK: Academic Press Ltd; 2002: 1045–1048.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

