Pervasive under-dominance in gene expression underlying emergent growth trajectories in Arabidopsis thaliana hybrids
- PMID: 37667232
- PMCID: PMC10478501
- DOI: 10.1186/s13059-023-03043-3
Pervasive under-dominance in gene expression underlying emergent growth trajectories in Arabidopsis thaliana hybrids
Abstract
Background: Complex traits, such as growth and fitness, are typically controlled by a very large number of variants, which can interact in both additive and non-additive fashion. In an attempt to gauge the relative importance of both types of genetic interactions, we turn to hybrids, which provide a facile means for creating many novel allele combinations.
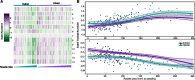
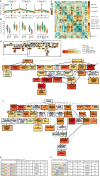
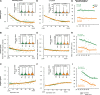
Results: We focus on the interaction between alleles of the same locus, i.e., dominance, and perform a transcriptomic study involving 141 random crosses between different accessions of the plant model species Arabidopsis thaliana. Additivity is rare, consistently observed for only about 300 genes enriched for roles in stress response and cell death. Regulatory rare-allele burden affects the expression level of these genes but does not correlate with F1 rosette size. Non-additive, dominant gene expression in F1 hybrids is much more common, with the vast majority of genes (over 90%) being expressed below the parental average. Unlike in the additive genes, regulatory rare-allele burden in the dominant gene set is strongly correlated with F1 rosette size, even though it only mildly covaries with the expression level of these genes.
Conclusions: Our study underscores under-dominance as the predominant gene action associated with emergence of rosette growth trajectories in the A. thaliana hybrid model. Our work lays the foundation for understanding molecular mechanisms and evolutionary forces that lead to dominance complementation of rare regulatory alleles.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
D.W. holds equity in Computomics, which advises breeders. The other authors declare that they have no competing interests.
Figures


References
-
- Falconer DS, Mackay TFC. Introduction to quantitative genetics. Essex: Longman; 1996.
-
- Braendle C, Heyland A, Flatt T. Integrating mechanistic and evolutionary analysis of life history variation. Mechanisms of life history evolution. The genetics and physiology of life history traits and trade-offs. New York: Oxford University Press; 2011. pp. 3–10.
-
- Mackay TFC, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–577. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

