Beyond rational-biosensor-guided isolation of 100 independently evolved bacterial strain variants and comparative analysis of their genomes
- PMID: 37667306
- PMCID: PMC10478468
- DOI: 10.1186/s12915-023-01688-x
Beyond rational-biosensor-guided isolation of 100 independently evolved bacterial strain variants and comparative analysis of their genomes
Abstract
Background: In contrast to modern rational metabolic engineering, classical strain development strongly relies on random mutagenesis and screening for the desired production phenotype. Nowadays, with the availability of biosensor-based FACS screening strategies, these random approaches are coming back into fashion. In this study, we employ this technology in combination with comparative genome analyses to identify novel mutations contributing to product formation in the genome of a Corynebacterium glutamicum L-histidine producer. Since all known genetic targets contributing to L-histidine production have been already rationally engineered in this strain, identification of novel beneficial mutations can be regarded as challenging, as they might not be intuitively linkable to L-histidine biosynthesis.
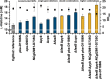
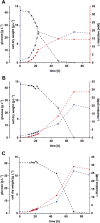
Results: In order to identify 100 improved strain variants that had each arisen independently, we performed > 600 chemical mutagenesis experiments, > 200 biosensor-based FACS screenings, isolated > 50,000 variants with increased fluorescence, and characterized > 4500 variants with regard to biomass formation and L-histidine production. Based on comparative genome analyses of these 100 variants accumulating 10-80% more L-histidine, we discovered several beneficial mutations. Combination of selected genetic modifications allowed for the construction of a strain variant characterized by a doubled L-histidine titer (29 mM) and product yield (0.13 C-mol C-mol-1) in comparison to the starting variant.
Conclusions: This study may serve as a blueprint for the identification of novel beneficial mutations in microbial producers in a more systematic manner. This way, also previously unexplored genes or genes with previously unknown contribution to the respective production phenotype can be identified. We believe that this technology has a great potential to push industrial production strains towards maximum performance.
Keywords: Biosensor; Comparative genome analysis; Corynebacterium glutamicum; Fluorescence-activated cell sorting; Genome sequencing; High-throughput screening; L-histidine; Metabolic engineering; Single-nucleotide polymorphism.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Lee SY, Kim HU. Systems strategies for developing industrial microbial strains. Nat Biotechnol. 2015;33(10):1061–1072. - PubMed
-
- Wehrs M, Tanjore D, Eng T, Lievense J, Pray TR, Mukhopadhyay A. Engineering robust production microbes for large-scale cultivation. Trends Microbiol. 2019;27(6):524–537. - PubMed
-
- Nielsen J, Keasling JD. Engineering cellular cetabolism. Cell. 2016;164(6):1185–1197. - PubMed
-
- Eggeling L, Bott M. A giant market and a powerful metabolism: l-lysine provided by Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2015;99(8):3387–3394. - PubMed
-
- Takors R, Bathe B, Rieping M, Hans S, Kelle R, Huthmacher K. Systems biology for industrial strains and fermentation processes—example: amino acids. J Biotechnol. 2007;129(2):181–190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

