Acute and Long-Term Consequences of Co-opted doublesex on the Development of Mimetic Butterfly Color Patterns
- PMID: 37668300
- PMCID: PMC10498343
- DOI: 10.1093/molbev/msad196
Acute and Long-Term Consequences of Co-opted doublesex on the Development of Mimetic Butterfly Color Patterns
Abstract
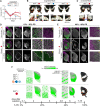
Novel phenotypes are increasingly recognized to have evolved by co-option of conserved genes into new developmental contexts, yet the process by which co-opted genes modify existing developmental programs remains obscure. Here, we provide insight into this process by characterizing the role of co-opted doublesex in butterfly wing color pattern development. dsx is the master regulator of insect sex differentiation but has been co-opted to control the switch between discrete nonmimetic and mimetic patterns in Papilio alphenor and its relatives through the evolution of novel mimetic alleles. We found dynamic spatial and temporal expression pattern differences between mimetic and nonmimetic butterflies throughout wing development. A mimetic color pattern program is switched on by a pulse of dsx expression in early pupal development that causes acute and long-term differential gene expression, particularly in Wnt and Hedgehog signaling pathways. RNAi suggested opposing, novel roles for these pathways in mimetic pattern development. Importantly, Dsx co-option caused Engrailed, a primary target of Hedgehog signaling, to gain a novel expression domain early in pupal wing development that is propagated through mid-pupal development to specify novel mimetic patterns despite becoming decoupled from Dsx expression itself. Altogether, our findings provide multiple views into how co-opted genes can both cause and elicit changes to conserved networks and pathways to result in development of novel, adaptive phenotypes.
Keywords: co-option; development; evolution; gene regulation; mimicry; polymorphism; sexual dimorphism.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



References
-
- Alexa A, Rahnenfuhrer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 22:1600–1607. - PubMed
-
- Ando T, Fujiwara H. 2013. Electroporation-mediated somatic transgenesis for rapid functional analysis in insects. Development. 140:454–458. - PubMed
-
- Banerjee TD, Ramos D, Monteiro A. 2020. Expression of multiple engrailed family genes in eyespots of Bicyclus anynana butterflies does not implicate the duplication events in the evolution of this morphological novelty. Front Ecol Evol. 8:227.
-
- Beldade P, Brakefield PM. 2002. The genetics and evo-devo of butterfly wing patterns. Nature. 3:442–452. - PubMed
-
- Brakefield PM, Gates J, Keys D, Kesbeke F, Wijngaarden PJ, Monteiro A, French V, Carroll SB. 1996. Development, plasticity and evolution of butterfly eyespot patterns. Nature. 384:236–242. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

