Integrated bioinformatics and validation reveal SOX12 as potential biomarker in colon adenocarcinoma based on an immune infiltration-related ceRNA network
- PMID: 37668799
- PMCID: PMC11797483
- DOI: 10.1007/s00432-023-05316-7
Integrated bioinformatics and validation reveal SOX12 as potential biomarker in colon adenocarcinoma based on an immune infiltration-related ceRNA network
Abstract
Purpose: The primary objective of this study was to construct competing endogenous RNA (ceRNA) networks and evaluate the prognostic significance of tumor-infiltrating immune cells (TIICs) and key biomarkers within the ceRNA networks in colon adenocarcinoma (COAD) patients.
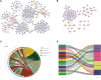
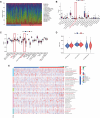
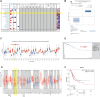
Methods: Comprehensive bioinformatics tools were used to screen differentially expressed genes (DEGs), miRNAs (DEMs), and lncRNAs (DELs) related to COAD, leading to the creation of ceRNA networks. The CIBERSORT technique was employed to assess the significance of TIICs in COAD, and an immune-related prognosis prediction model was subsequently developed. Co-expression analyses were conducted to determine the relationship between key genes in ceRNA networks and immunologically significant TIICs. The study also utilized 5 GEO datasets and web-based databases to externally validate the findings.
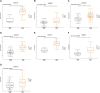
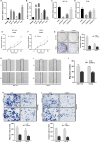
Results: The study revealed a statistically significant relationship between key hub genes and immune cells, as determined through co-expression analysis. Two hub regulators (SOX12 and H19) demonstrated significant prognostic value in the ceRNA-related prognostic model, and their elevated expression levels were verified across multiple CRC cell lines. Additionally, the knockdown of SOX12 led to a suppression of proliferation, migration, and invasion in colon cancer cells.
Conclusion: Through the construction of ceRNA networks and evaluation of TIICs, the study successfully established two risk score models and nomograms. These models serve as valuable tools for understanding the molecular processes and predicting the prognosis of COAD patients. Further validation of hub regulators SOX12 and H19 substantiates their potential role as key biomarkers in COAD.
Keywords: Colon adenocarcinoma; Immune infiltration; Nomogram; Prognostic model; ceRNA network.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, Soong DYH, Cotechini T, Anur P, Lin EY, Fidanza A, Lopez-Yrigoyen M, Millar MR, Urman A, Ai Z, Spellman PT, Hwang ES, Dixon JM, Wiechmann L, Coussens LM, Smith HO, Pollard JW (2019) Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 35:588 e510-602 e510 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

