Chloroplast import motor subunits FtsHi1 and FtsHi2 are located on opposite sides of the inner envelope membrane
- PMID: 37669373
- PMCID: PMC10500165
- DOI: 10.1073/pnas.2307747120
Chloroplast import motor subunits FtsHi1 and FtsHi2 are located on opposite sides of the inner envelope membrane
Abstract
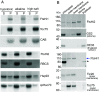
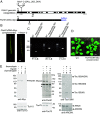
Protein import into chloroplasts is powered by ATP hydrolysis in the stroma. Establishing the identity and functional mechanism of the stromal ATPase motor that drives import is critical for understanding chloroplast biogenesis. Recently, a complex consisting of Ycf2, FtsHi1, FtsHi2, FtsHi4, FtsHi5, FtsH12, and malate dehydrogenase was shown to be important for chloroplast protein import, and it has been proposed to act as the motor driving protein translocation across the chloroplast envelope into the stroma. To gain further mechanistic understanding of how the motor functions, we performed membrane association and topology analyses on two of its subunits, FtsHi1 and FtsHi2. We isolated cDNA clones encoding FtsHi1 and FtsHi2 preproteins to perform in vitro import experiments in order to determine the exact size of each mature protein. We also generated antibodies against the C-termini of the proteins, i.e., where their ATPase domains reside. Protease treatments and alkaline and high-salt extractions of chloroplasts with imported and endogenous proteins revealed that FtsHi1 is an integral membrane protein with its C-terminal portion located in the intermembrane space of the envelope, not the stroma, whereas FtsHi2 is a soluble protein in the stroma. We further complemented an FtsHi1-knockout mutant with a C-terminally tagged FtsHi1 and obtained identical results for topological analyses. Our data indicate that the model of a single membrane-anchored pulling motor at the stromal side of the inner membrane needs to be revised and suggest that the Ycf2-FtsHi complex may have additional functions.
Keywords: chloroplast; protein import; protein topology.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
A Ycf2-FtsHi Heteromeric AAA-ATPase Complex Is Required for Chloroplast Protein Import.Plant Cell. 2018 Nov;30(11):2677-2703. doi: 10.1105/tpc.18.00357. Epub 2018 Oct 11. Plant Cell. 2018. PMID: 30309901 Free PMC article.
-
Conservation and specialization of the Ycf2-FtsHi chloroplast protein import motor in green algae.Cell. 2024 Oct 3;187(20):5638-5650.e18. doi: 10.1016/j.cell.2024.08.002. Epub 2024 Aug 27. Cell. 2024. PMID: 39197449
-
Abundance of metalloprotease FtsH12 modulates chloroplast development in Arabidopsis thaliana.J Exp Bot. 2021 Apr 13;72(9):3455-3473. doi: 10.1093/jxb/eraa550. J Exp Bot. 2021. PMID: 33216923 Free PMC article.
-
Molecular chaperone involvement in chloroplast protein import.Biochim Biophys Acta. 2013 Feb;1833(2):332-40. doi: 10.1016/j.bbamcr.2012.03.019. Epub 2012 Apr 12. Biochim Biophys Acta. 2013. PMID: 22521451 Review.
-
Protein import into cyanelles and complex chloroplasts.Plant Mol Biol. 1998 Sep;38(1-2):247-63. Plant Mol Biol. 1998. PMID: 9738970 Review.
References
-
- Sun Y., Jarvis R. P., Chloroplast proteostasis: Import, sorting, ubiquitination, and proteolysis. Annu. Rev. Plant Biol. 74, 259–283 (2023). - PubMed
-
- Theg S. M., Bauerle C., Olsen L. J., Selman B. R., Keegstra K., Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J. Biol. Chem. 264, 6730–6736 (1989). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

