Mouse models of SYNGAP1-related intellectual disability
- PMID: 37669379
- PMCID: PMC10500186
- DOI: 10.1073/pnas.2308891120
Mouse models of SYNGAP1-related intellectual disability
Abstract
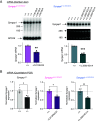
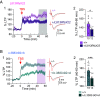
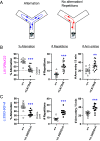
SYNGAP1 is a Ras-GTPase-activating protein highly enriched at excitatory synapses in the brain. De novo loss-of-function mutations in SYNGAP1 are a major cause of genetically defined neurodevelopmental disorders (NDDs). These mutations are highly penetrant and cause SYNGAP1-related intellectual disability (SRID), an NDD characterized by cognitive impairment, social deficits, early-onset seizures, and sleep disturbances. Studies in rodent neurons have shown that Syngap1 regulates developing excitatory synapse structure and function, and heterozygous Syngap1 knockout mice have deficits in synaptic plasticity, learning, and memory and have seizures. However, how specific SYNGAP1 mutations found in humans lead to disease has not been investigated in vivo. To explore this, we utilized the CRISPR-Cas9 system to generate knock-in mouse models with two distinct known causal variants of SRID: one with a frameshift mutation leading to a premature stop codon, SYNGAP1; L813RfsX22, and a second with a single-nucleotide mutation in an intron that creates a cryptic splice acceptor site leading to premature stop codon, SYNGAP1; c.3583-9G>A. While reduction in Syngap1 mRNA varies from 30 to 50% depending on the specific mutation, both models show ~50% reduction in Syngap1 protein, have deficits in synaptic plasticity, and recapitulate key features of SRID including hyperactivity and impaired working memory. These data suggest that half the amount of SYNGAP1 protein is key to the pathogenesis of SRID. These results provide a resource to study SRID and establish a framework for the development of therapeutic strategies for this disorder.
Keywords: dendritic development; neurodevelopmental disorders; synaptic GTPase-activating protein; synaptic plasticity.
Conflict of interest statement
The authors declare no competing interest.
Figures



Update of
-
Mouse models of SYNGAP1 -related intellectual disability.bioRxiv [Preprint]. 2023 May 26:2023.05.25.542312. doi: 10.1101/2023.05.25.542312. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Sep 12;120(37):e2308891120. doi: 10.1073/pnas.2308891120. PMID: 37293116 Free PMC article. Updated. Preprint.
References
-
- Fischbach G. D., Lord C., The Simons Simplex Collection: A resource for identification of autism genetic risk factors. Neuron 68, 192–195 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

