Notch and retinoic acid signals regulate macrophage formation from endocardium downstream of Nkx2-5
- PMID: 37669937
- PMCID: PMC10480477
- DOI: 10.1038/s41467-023-41039-6
Notch and retinoic acid signals regulate macrophage formation from endocardium downstream of Nkx2-5
Abstract
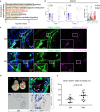
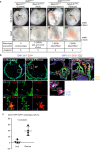
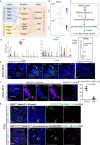
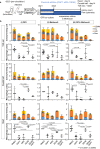
Hematopoietic progenitors are enriched in the endocardial cushion and contribute, in a Nkx2-5-dependent manner, to tissue macrophages required for the remodeling of cardiac valves and septa. However, little is known about the molecular mechanism of endocardial-hematopoietic transition. In the current study, we identified the regulatory network of endocardial hematopoiesis. Signal network analysis from scRNA-seq datasets revealed that genes in Notch and retinoic acid (RA) signaling are significantly downregulated in Nkx2-5-null endocardial cells. In vivo and ex vivo analyses validate that the Nkx2-5-Notch axis is essential for the generation of both hemogenic and cushion endocardial cells, and the suppression of RA signaling via Dhrs3 expression plays important roles in further differentiation into macrophages. Genetic ablation study revealed that these macrophages are essential in cardiac valve remodeling. In summary, the study demonstrates that the Nkx2-5/Notch/RA signaling plays a pivotal role in macrophage differentiation from hematopoietic progenitors.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

