Demonstrating paths for unlocking the value of cloud genomics through cross cohort analysis
- PMID: 37669985
- PMCID: PMC10480504
- DOI: 10.1038/s41467-023-41185-x
Demonstrating paths for unlocking the value of cloud genomics through cross cohort analysis
Abstract
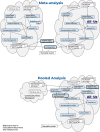
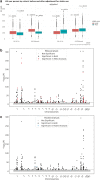
Recently, large scale genomic projects such as All of Us and the UK Biobank have introduced a new research paradigm where data are stored centrally in cloud-based Trusted Research Environments (TREs). To characterize the advantages and drawbacks of different TRE attributes in facilitating cross-cohort analysis, we conduct a Genome-Wide Association Study of standard lipid measures using two approaches: meta-analysis and pooled analysis. Comparison of full summary data from both approaches with an external study shows strong correlation of known loci with lipid levels (R2 ~ 83-97%). Importantly, 90 variants meet the significance threshold only in the meta-analysis and 64 variants are significant only in pooled analysis, with approximately 20% of variants in each of those groups being most prevalent in non-European, non-Asian ancestry individuals. These findings have important implications, as technical and policy choices lead to cross-cohort analyses generating similar, but not identical results, particularly for non-European ancestral populations.
© 2023. Springer Nature Limited.
Conflict of interest statement
P.N. reports investigator-initiated grants from Amgen, Apple, AstraZeneca, Boston Scientific, and Novartis, personal fees from Apple, AstraZeneca, Blackstone Life Sciences, Foresite Labs, Novartis, Roche/Genentech, is a co-founder of TenSixteen Bio, is a shareholder of geneXwell and TenSixteen Bio, and spousal employment at Vertex, all unrelated to the present work. A.G.B. is a co-founder and shareholder of TenSixteen Bio unrelated to the present work. N.D. and D.G. are employees of Verily Life Sciences and may own stock as part of the standard compensation package. A.P. serves as a Google Ventures (GV) venture partner and holds an equity interest in certain of GV’s affiliated investment funds. A.P. has also received funding from Verily, MSFT, Intel, IBM, Bayer, Pfizer, Astra Zeneca, and Biogen. The remaining authors declare no competing interests.
Figures


References
-
- UK Health Data Research Alliance & NHSX. Building Trusted Research Environments - principles and best practices; Towards TRE ecosystems. Preprint at 10.5281/ZENODO.5767586 (2021).
-
- Hubbard, T., Reilly, G., Varma, S. & Seymour, D. Trusted research environments (TRE) green paper. Preprint at 10.5281/ZENODO.4594704 (2020).
Publication types
MeSH terms
Substances
Grants and funding
- U2C OD023196/OD/NIH HHS/United States
- OT2 OD026551/OD/NIH HHS/United States
- OT2 OD026552/OD/NIH HHS/United States
- OT2 OD026549/OD/NIH HHS/United States
- OT2 OD025337/OD/NIH HHS/United States
- R01 HL142711/HL/NHLBI NIH HHS/United States
- OT2 OD025277/OD/NIH HHS/United States
- OT2 OD026550/OD/NIH HHS/United States
- OT2 OD026553/OD/NIH HHS/United States
- OT2 OD023205/OD/NIH HHS/United States
- OT2 OD025276/OD/NIH HHS/United States
- OT2 OD026554/OD/NIH HHS/United States
- U24 OD023163/OD/NIH HHS/United States
- OT2 OD023206/OD/NIH HHS/United States
- OT2 OD002748/OD/NIH HHS/United States
- DP5 OD029586/OD/NIH HHS/United States
- OT2 OD026556/OD/NIH HHS/United States
- U24 OD023176/OD/NIH HHS/United States
- OT2 OD026548/OD/NIH HHS/United States
- U01 HG011719/HG/NHGRI NIH HHS/United States
- OT2 OD025315/OD/NIH HHS/United States
- U24 OD023121/OD/NIH HHS/United States
- R01 HL127564/HL/NHLBI NIH HHS/United States
- OT2 OD002750/OD/NIH HHS/United States
- OT2 OD026555/OD/NIH HHS/United States
- OT2 OD026557/OD/NIH HHS/United States
- OT2 OD002751/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources

