Genomic analysis of Chlamydia psittaci from a multistate zoonotic outbreak in two chicken processing plants
- PMID: 37670735
- PMCID: PMC10475345
- DOI: 10.7150/jgen.86558
Genomic analysis of Chlamydia psittaci from a multistate zoonotic outbreak in two chicken processing plants
Abstract
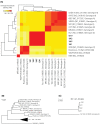
Four Chlamydia psittaci isolates were recovered from clinical specimens from ill workers during a multistate outbreak at two chicken processing plants. Whole genome sequencing analyses revealed high similarity to C. psittaci genotype D. The isolates differed from each other by only two single nucleotide polymorphisms, indicating a common source.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
References
-
- AbdelRahman YM, Belland RJ. The Chlamydial Developmental Cycle. FEMS Microbiology Reviews. 2005;29:949–59. - PubMed
-
- Hotzel H, Berndt A, Melzer F, Sachse K. Occurrence of Chlamydiaceae spp. in a wild boar (Sus scrofa L.) population in Thuringia (Germany) Veterinary Microbiology. 2004;103:121–6. - PubMed
-
- Balsamo G, Maxted AM, Midla JW, Murphy JM, Wohrle R, Edling TM. et al. Compendium of Measures to Control Chlamydia psittaci Infection Among Humans (Psittacosis) and Pet Birds (Avian Chlamydiosis), 2017. Journal of Avian Medicine and Surgery. 2017;31:262–82. - PubMed
LinkOut - more resources
Full Text Sources

