Impact of conservation tillage on wheat performance and its microbiome
- PMID: 37670872
- PMCID: PMC10475739
- DOI: 10.3389/fpls.2023.1211758
Impact of conservation tillage on wheat performance and its microbiome
Abstract
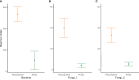
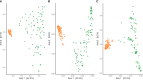
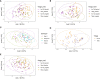
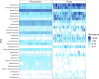
Winter wheat is an important cereal consumed worldwide. However, current management practices involving chemical fertilizers, irrigation, and intensive tillage may have negative impacts on the environment. Conservation agriculture is often presented as a sustainable alternative to maintain wheat production, favoring the beneficial microbiome. Here, we evaluated the impact of different water regimes (rainfed and irrigated), fertilization levels (half and full fertilization), and tillage practices (occasional tillage and no-tillage) on wheat performance, microbial activity, and rhizosphere- and root-associated microbial communities of four winter wheat genotypes (Antequera, Allez-y, Apache, and Cellule) grown in a field experiment. Wheat performance (i.e., yield, plant nitrogen concentrations, and total nitrogen uptake) was mainly affected by irrigation, fertilization, and genotype, whereas microbial activity (i.e., protease and alkaline phosphatase activities) was affected by irrigation. Amplicon sequencing data revealed that habitat (rhizosphere vs. root) was the main factor shaping microbial communities and confirmed that the selection of endophytic microbial communities takes place thanks to specific plant-microbiome interactions. Among the experimental factors applied, the interaction of irrigation and tillage influenced rhizosphere- and root-associated microbiomes. The findings presented in this work make it possible to link agricultural practices to microbial communities, paving the way for better monitoring of these microorganisms in the context of agroecosystem sustainability.
Keywords: amplicon sequencing; fertilization; irrigation; soil microbiome; tillage; wheat genotype.
Copyright © 2023 Romano, Bodenhausen, Basch, Soares, Faist, Trognitz, Sessitsch, Doubell, Declerck and Symanczik.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Long-term conservation tillage with reduced nitrogen fertilization intensity can improve winter wheat health via positive plant-microorganism feedback in the rhizosphere.FEMS Microbiol Ecol. 2024 Jan 24;100(2):fiae003. doi: 10.1093/femsec/fiae003. FEMS Microbiol Ecol. 2024. PMID: 38224956 Free PMC article.
-
Integrated organic and inorganic fertilization and reduced irrigation altered prokaryotic microbial community and diversity in different compartments of wheat root zone contributing to improved nitrogen uptake and wheat yield.Sci Total Environ. 2022 Oct 10;842:156952. doi: 10.1016/j.scitotenv.2022.156952. Epub 2022 Jun 22. Sci Total Environ. 2022. PMID: 35752240
-
Influence of Fertilization Methods and Types on Wheat Rhizosphere Microbiome Community and Functions.J Agric Food Chem. 2024 Apr 10;72(14):7794-7806. doi: 10.1021/acs.jafc.3c09941. Epub 2024 Apr 1. J Agric Food Chem. 2024. PMID: 38561246
-
Effects of Fallow Management Practices on Soil Water, Crop Yield and Water Use Efficiency in Winter Wheat Monoculture System: A Meta-Analysis.Front Plant Sci. 2022 Apr 27;13:825309. doi: 10.3389/fpls.2022.825309. eCollection 2022. Front Plant Sci. 2022. PMID: 35574095 Free PMC article.
-
Towards sustainable agriculture: rhizosphere microbiome engineering.Appl Microbiol Biotechnol. 2021 Oct;105(19):7141-7160. doi: 10.1007/s00253-021-11555-w. Epub 2021 Sep 11. Appl Microbiol Biotechnol. 2021. PMID: 34508284 Review.
Cited by
-
Management of soil cover and tillage regimes in upland rice-sweet corn systems for better system performance, energy use and carbon footprints.Heliyon. 2024 Feb 15;10(4):e26524. doi: 10.1016/j.heliyon.2024.e26524. eCollection 2024 Feb 29. Heliyon. 2024. PMID: 38420378 Free PMC article.
-
Inoculation with a microbial consortium increases soil microbial diversity and improves agronomic traits of tomato under water and nitrogen deficiency.Front Plant Sci. 2023 Dec 6;14:1304627. doi: 10.3389/fpls.2023.1304627. eCollection 2023. Front Plant Sci. 2023. PMID: 38126011 Free PMC article.
-
Long-term conservation tillage with reduced nitrogen fertilization intensity can improve winter wheat health via positive plant-microorganism feedback in the rhizosphere.FEMS Microbiol Ecol. 2024 Jan 24;100(2):fiae003. doi: 10.1093/femsec/fiae003. FEMS Microbiol Ecol. 2024. PMID: 38224956 Free PMC article.
References
-
- Azaroual S. E., Kasmi Y., Aasfar A., El Arroussi H., Zeroual Y., El Kadiri Y., et al. . (2022). Investigation of bacterial diversity using 16S rRNA sequencing and prediction of its functionalities in Moroccan phosphate mine ecosystem. Sci. Rep. 12, 3741. doi: 10.1038/s41598-022-07765-5 - DOI - PMC - PubMed
-
- Bakker M. G., Manter D. K., Sheflin A. M., Weir T. L., Vivanco J. M. (2012). Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 360, 1–13. doi: 10.1007/s11104-012-1361-x - DOI
LinkOut - more resources
Full Text Sources

