Synthetic whole-slide image tile generation with gene expression profile-infused deep generative models
- PMID: 37671024
- PMCID: PMC10475789
- DOI: 10.1016/j.crmeth.2023.100534
Synthetic whole-slide image tile generation with gene expression profile-infused deep generative models
Abstract
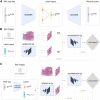
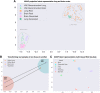
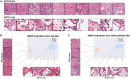
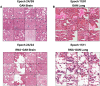
In this work, we propose an approach to generate whole-slide image (WSI) tiles by using deep generative models infused with matched gene expression profiles. First, we train a variational autoencoder (VAE) that learns a latent, lower-dimensional representation of multi-tissue gene expression profiles. Then, we use this representation to infuse generative adversarial networks (GANs) that generate lung and brain cortex tissue tiles, resulting in a new model that we call RNA-GAN. Tiles generated by RNA-GAN were preferred by expert pathologists compared with tiles generated using traditional GANs, and in addition, RNA-GAN needs fewer training epochs to generate high-quality tiles. Finally, RNA-GAN was able to generalize to gene expression profiles outside of the training set, showing imputation capabilities. A web-based quiz is available for users to play a game distinguishing real and synthetic tiles: https://rna-gan.stanford.edu/, and the code for RNA-GAN is available here: https://github.com/gevaertlab/RNA-GAN.
Keywords: artificial intelligence; deep learning; generative adversarial network; generative model; synthetic biomedical data; variational autoencoder.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Coudray N., Ocampo P.S., Sakellaropoulos T., Narula N., Snuderl M., Fenyö D., Moreira A.L., Razavian N., Tsirigos A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

