Pan-conserved segment tags identify ultra-conserved sequences across assemblies in the human pangenome
- PMID: 37671027
- PMCID: PMC10475782
- DOI: 10.1016/j.crmeth.2023.100543
Pan-conserved segment tags identify ultra-conserved sequences across assemblies in the human pangenome
Abstract
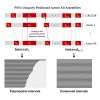
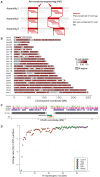
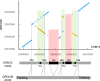
The human pangenome, a new reference sequence, addresses many limitations of the current GRCh38 reference. The first release is based on 94 high-quality haploid assemblies from individuals with diverse backgrounds. We employed a k-mer indexing strategy for comparative analysis across multiple assemblies, including the pangenome reference, GRCh38, and CHM13, a telomere-to-telomere reference assembly. Our k-mer indexing approach enabled us to identify a valuable collection of universally conserved sequences across all assemblies, referred to as "pan-conserved segment tags" (PSTs). By examining intervals between these segments, we discerned highly conserved genomic segments and those with structurally related polymorphisms. We found 60,764 polymorphic intervals with unique geo-ethnic features in the pangenome reference. In this study, we utilized ultra-conserved sequences (PSTs) to forge a link between human pangenome assemblies and reference genomes. This methodology enables the examination of any sequence of interest within the pangenome, using the reference genome as a comparative framework.
Keywords: k-mer; pan-conserved segment; pangenome; reference genome; structural polymorphism; structural variations.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Zhou B., Arthur J.G., Guo H., Hughes C.R., Kim T., Huang Y., Pattni R., Lee H., Ji H.P., Song G., et al. Automatic detection of complex structural genome variation across world populations. bioRxiv. 2023 doi: 10.1101/200170. Preprint at. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

