Shigella induces epigenetic reprogramming of zebrafish neutrophils
- PMID: 37672585
- PMCID: PMC10482349
- DOI: 10.1126/sciadv.adf9706
Shigella induces epigenetic reprogramming of zebrafish neutrophils
Abstract
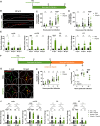
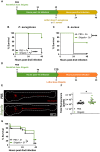
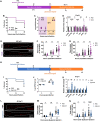
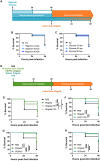
Trained immunity is a long-term memory of innate immune cells, generating an improved response upon reinfection. Shigella is an important human pathogen and inflammatory paradigm for which there is no effective vaccine. Using zebrafish larvae, we demonstrate that after Shigella training, neutrophils are more efficient at bacterial clearance. We observe that Shigella-induced protection is nonspecific and has differences with training by BCG and β-glucan. Analysis of histone ChIP-seq on trained neutrophils revealed that Shigella training deposits the active H3K4me3 mark on promoter regions of 1612 genes, dramatically changing the epigenetic landscape of neutrophils toward enhanced microbial recognition and mitochondrial ROS production. Last, we demonstrate that mitochondrial ROS plays a key role in enhanced antimicrobial activity of trained neutrophils. It is envisioned that signals and mechanisms we discover here can be used in other vertebrates, including humans, to suggest new therapeutic strategies involving neutrophils to control bacterial infection.
Figures

References
-
- J. Domínguez-Andrés, J. C. Dos Santos, S. Bekkering, W. J. M. Mulder, J. W. M. van der Meer, N. P. Riksen, L. A. B. Joosten, M. G. Netea, Trained immunity: Adaptation within innate immune mechanisms. Physiol. Rev. 103, 313–346 (2023). - PubMed
-
- R. J. W. Arts, A. Carvalho, C. La Rocca, C. Palma, F. Rodrigues, R. Silvestre, J. Kleinnijenhuis, E. Lachmandas, L. G. Gonçalves, A. Belinha, C. Cunha, M. Oosting, L. A. B. Joosten, G. Matarese, R. van Crevel, M. G. Netea, Immunometabolic pathways in BCG-induced trained immunity. Cell Rep. 17, 2562–2571 (2016). - PMC - PubMed
-
- E. Kaufmann, J. Sanz, J. L. Dunn, N. Khan, L. E. Mendonça, A. Pacis, F. Tzelepis, E. Pernet, A. Dumaine, J. C. Grenier, F. Mailhot-Léonard, E. Ahmed, J. Belle, R. Besla, B. Mazer, I. L. King, A. Nijnik, C. S. Robbins, L. B. Barreiro, M. Divangahi, BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell 172, 176–190.e19 (2018). - PubMed
-
- B. de Laval, J. Maurizio, P. K. Kandalla, G. Brisou, L. Simonnet, C. Huber, G. Gimenez, O. Matcovitch-Natan, S. Reinhardt, E. David, A. Mildner, A. Leutz, B. Nadel, C. Bordi, I. Amit, S. Srrazin, M. H. Sieweke, C/EBPβ-dependent epigenetic memory induces trained immunity in hematopoietic stem cells. Cell Stem Cell 26, 657–674.e8 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

