The RasGAP-associated endoribonuclease G3BP mediates stress granule assembly
- PMID: 37672657
- PMCID: PMC10482220
- DOI: 10.1083/jcb.200212128072023new
The RasGAP-associated endoribonuclease G3BP mediates stress granule assembly
Abstract
Stress granules (SGs) are formed in the cytoplasm in response to various toxic agents and are believed to play a critical role in the regulation of mRNA metabolism during stress. In SGs, mRNAs are stored in an abortive translation initiation complex that can be routed to either translation initiation or degradation. Here, we show that G3BP, a phosphorylation-dependent endoribonuclease that interacts with RasGAP, is recruited to SGs in cells exposed to arsenite. G3BP may thus determine the fate of mRNAs during cellular stress. Remarkably, SG assembly can be either dominantly induced by G3BP overexpression, or on the contrary, inhibited by expressing a central domain of G3BP. This region binds RasGAP and contains serine 149 whose dephosphorylation is induced by arsenite treatment. Critically, a non-phosphorylatable G3BP mutant (S149A) oligomerizes and assembles SG. These results suggest that G3BP is an effector of SG assembly and that Ras signaling contributes to this process by regulating G3BP dephosphorylation.
© 2023 Tourrière et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures
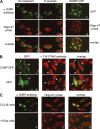
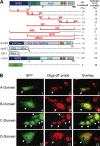
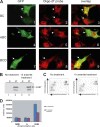
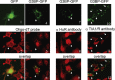

Corrected and republished from
- doi: 10.1083/jcb.200212128
References
-
- Allemand, E., Gattoni R., Bourbon H.M., Stevenin J., Cáceres J.F., Soret J., and Tazi J.. 2001. Distinctive features of Drosophila alternative splicing factor RS domain: Implication for specific phosphorylation, shuttling, and splicing activation. Mol. Cell. Biol. 21:1345–1359. 10.1128/MCB.21.4.1345-1359.2001 - DOI - PMC - PubMed
-
- Allemand, E., Dokudovskaya S., Bordonné R., and Tazi J.. 2002. A conserved Drosophila transportin-serine/arginine-rich (SR) protein permits nuclear import of Drosophila SR protein splicing factors and their antagonist repressor splicing factor 1. Mol. Biol. Cell. 13:2436–2447. 10.1091/mbc.e02-02-0102 - DOI - PMC - PubMed
-
- Gallouzi, I.E., Parker F., Chebli K., Maurier F., Labourier E., Barlat I., Capony J.P., Tocque B., and Tazi J.. 1998. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: A potential link between signal transduction and RNA stability. Mol. Cell. Biol. 18:3956–3965. 10.1128/MCB.18.7.3956 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

