Complete human day 14 post-implantation embryo models from naive ES cells
- PMID: 37673118
- PMCID: PMC10584686
- DOI: 10.1038/s41586-023-06604-5
Complete human day 14 post-implantation embryo models from naive ES cells
Abstract
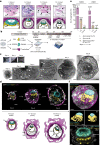
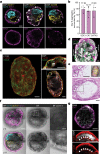
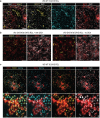
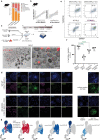
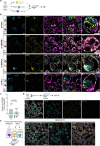
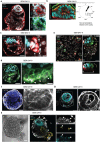
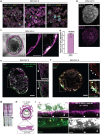
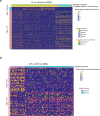
The ability to study human post-implantation development remains limited owing to ethical and technical challenges associated with intrauterine development after implantation1. Embryo-like models with spatially organized morphogenesis and structure of all defining embryonic and extra-embryonic tissues of the post-implantation human conceptus (that is, the embryonic disc, the bilaminar disc, the yolk sac, the chorionic sac and the surrounding trophoblast layer) remain lacking1,2. Mouse naive embryonic stem cells have recently been shown to give rise to embryonic and extra-embryonic stem cells capable of self-assembling into post-gastrulation structured stem-cell-based embryo models with spatially organized morphogenesis (called SEMs)3. Here we extend those findings to humans using only genetically unmodified human naive embryonic stem cells (cultured in human enhanced naive stem cell medium conditions)4. Such human fully integrated and complete SEMs recapitulate the organization of nearly all known lineages and compartments of post-implantation human embryos, including the epiblast, the hypoblast, the extra-embryonic mesoderm and the trophoblast layer surrounding the latter compartments. These human complete SEMs demonstrated developmental growth dynamics that resemble key hallmarks of post-implantation stage embryogenesis up to 13-14 days after fertilization (Carnegie stage 6a). These include embryonic disc and bilaminar disc formation, epiblast lumenogenesis, polarized amniogenesis, anterior-posterior symmetry breaking, primordial germ-cell specification, polarized yolk sac with visceral and parietal endoderm formation, extra-embryonic mesoderm expansion that defines a chorionic cavity and a connecting stalk, and a trophoblast-surrounding compartment demonstrating syncytium and lacunae formation. This SEM platform will probably enable the experimental investigation of previously inaccessible windows of human early post implantation up to peri-gastrulation development.
© 2023. The Author(s).
Conflict of interest statement
J.H.H. together with B.O., E.W., V.B., A.A.-C., N.N., S.V., S.T., C.J., T.S. and F.R. submitted (through Yeda–Weizmann Institute of Science) patent applications relevant to the findings and technologies reported and media compositions utilized herein: (1) “Isolated naive pluripotent stem cells and methods of generating same” filed on 23 April 2013 (EP 61/814,920); (2) “Media for culturing naive pluripotent stem cells” filed on 30 July 2014 (EP 62/030,792); (3) “Culture media for pluripotent stem cells” filed on 23 January 2020 (EP 62/795,626); (4) “Methods and devices for ex-utero mouse embryonic development” (for natural and synthetic embryos) filed on 16 March 2021 (EP 281,561); (5) “Methods of generating a synthetic embryo” (for mouse and human SEMs/synthetic embryo models) filed on 6 March 2022 (EP 63/317,036). J.H.H. is a co-founder and chief scientific advisor of Renewal Bio, which has licensed technologies described herein and co-funded some parts of this project. RSeT defined human naive-like pluripotency growth medium was licensed and commercialized by Stem Cell Technologies based on some of the indicated patents/patent applications above (RSeT is the commercialized version of NHSM growth medium originally generated and patented by J.H.H. and N.N). The remaining authors declare no competing interests.
Figures












Comment in
-
Stem cells used to model a two-week-old human embryo.Nature. 2023 Oct;622(7983):469-470. doi: 10.1038/d41586-023-03150-y. Nature. 2023. PMID: 37848524 No abstract available.
-
A new advanced stem cell-based embryonic model: The ultimate model?Mol Ther. 2023 Nov 1;31(11):3109-3110. doi: 10.1016/j.ymthe.2023.10.013. Epub 2023 Oct 21. Mol Ther. 2023. PMID: 37866361 Free PMC article. No abstract available.
References
-
- Oldak, B. et al. Transgene-free ex utero derivation of a human post-implantation embryo model solely from genetically unmodified naive PSCs. Preprint at bioRxiv10.1101/2023.06.14.544922 (2023).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

