Circadian rhythm disruptions associated with opioid use disorder in synaptic proteomes of human dorsolateral prefrontal cortex and nucleus accumbens
- PMID: 37674018
- PMCID: PMC10914630
- DOI: 10.1038/s41380-023-02241-6
Circadian rhythm disruptions associated with opioid use disorder in synaptic proteomes of human dorsolateral prefrontal cortex and nucleus accumbens
Abstract
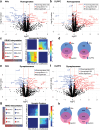
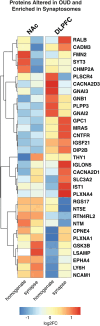
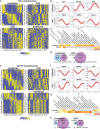
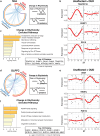
Opioid craving and relapse vulnerability is associated with severe and persistent sleep and circadian rhythm disruptions. Understanding the neurobiological underpinnings of circadian rhythms and opioid use disorder (OUD) may prove valuable for developing new treatments for opioid addiction. Previous work indicated molecular rhythm disruptions in the human brain associated with OUD, highlighting synaptic alterations in the dorsolateral prefrontal cortex (DLPFC) and nucleus accumbens (NAc)-key brain regions involved in cognition and reward, and heavily implicated in the pathophysiology of OUD. To provide further insights into the synaptic alterations in OUD, we used mass-spectrometry based proteomics to deeply profile protein expression alterations in bulk tissue and synaptosome preparations from DLPFC and NAc of unaffected and OUD subjects. We identified 55 differentially expressed (DE) proteins in DLPFC homogenates, and 44 DE proteins in NAc homogenates, between unaffected and OUD subjects. In synaptosomes, we identified 161 and 56 DE proteins in DLPFC and NAc, respectively, of OUD subjects. By comparing homogenate and synaptosome protein expression, we identified proteins enriched specifically in synapses that were significantly altered in both DLPFC and NAc of OUD subjects. Across brain regions, synaptic protein alterations in OUD subjects were primarily identified in glutamate, GABA, and circadian rhythm signaling. Using time-of-death (TOD) analyses, where the TOD of each subject is used as a time-point across a 24-h cycle, we were able to map circadian-related changes associated with OUD in synaptic proteomes associated with vesicle-mediated transport and membrane trafficking in the NAc and platelet-derived growth factor receptor beta signaling in DLPFC. Collectively, our findings lend further support for molecular rhythm disruptions in synaptic signaling in the human brain as a key factor in opioid addiction.
© 2023. The Author(s).
Conflict of interest statement
The authors report no competing interests.
Figures

Update of
-
Circadian rhythm disruptions associated with opioid use disorder in the synaptic proteomes of the human dorsolateral prefrontal cortex and nucleus accumbens.bioRxiv [Preprint]. 2023 Aug 24:2023.04.07.536056. doi: 10.1101/2023.04.07.536056. bioRxiv. 2023. Update in: Mol Psychiatry. 2023 Nov;28(11):4777-4792. doi: 10.1038/s41380-023-02241-6. PMID: 37066169 Free PMC article. Updated. Preprint.
References
-
- National Institute on Drug Abuse. Drug overdose death rates. National Institute on Drug Abuse; 2023. https://nida.nih.gov/research-topics/trends-statistics/overdose-death-rates. Accessed 23 March 2023.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

