Polθ is phosphorylated by PLK1 to repair double-strand breaks in mitosis
- PMID: 37674080
- PMCID: PMC10499603
- DOI: 10.1038/s41586-023-06506-6
Polθ is phosphorylated by PLK1 to repair double-strand breaks in mitosis
Erratum in
-
Publisher Correction: Polθ is phosphorylated by PLK1 to repair double-strand breaks in mitosis.Nature. 2024 Feb;626(7999):E13. doi: 10.1038/s41586-024-07025-8. Nature. 2024. PMID: 38273133 Free PMC article. No abstract available.
Abstract
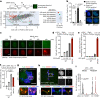
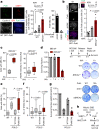
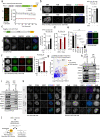
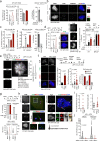
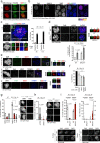
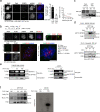
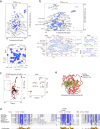
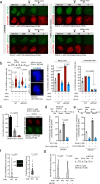
DNA double-strand breaks (DSBs) are deleterious lesions that challenge genome integrity. To mitigate this threat, human cells rely on the activity of multiple DNA repair machineries that are tightly regulated throughout the cell cycle1. In interphase, DSBs are mainly repaired by non-homologous end joining and homologous recombination2. However, these pathways are completely inhibited in mitosis3-5, leaving the fate of mitotic DSBs unknown. Here we show that DNA polymerase theta6 (Polθ) repairs mitotic DSBs and thereby maintains genome integrity. In contrast to other DSB repair factors, Polθ function is activated in mitosis upon phosphorylation by Polo-like kinase 1 (PLK1). Phosphorylated Polθ is recruited by a direct interaction with the BRCA1 C-terminal domains of TOPBP1 to mitotic DSBs, where it mediates joining of broken DNA ends. Loss of Polθ leads to defective repair of mitotic DSBs, resulting in a loss of genome integrity. This is further exacerbated in cells that are deficient in homologous recombination, where loss of mitotic DSB repair by Polθ results in cell death. Our results identify mitotic DSB repair as the underlying cause of synthetic lethality between Polθ and homologous recombination. Together, our findings reveal the critical importance of mitotic DSB repair in the maintenance of genome integrity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

