A novel natural killer-related signature to effectively predict prognosis in hepatocellular carcinoma
- PMID: 37674210
- PMCID: PMC10481539
- DOI: 10.1186/s12920-023-01638-0
A novel natural killer-related signature to effectively predict prognosis in hepatocellular carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is a prevalent tumor that poses a significant threat to human health, with 80% of cases being primary HCC. At present, Early diagnosis and predict prognosis of HCC is challenging and the it is characterized by a high degree of invasiveness, both of which negatively impact patient prognosis. Natural killer cells (NK) play an important role in the development, diagnosis and prognosis of malignant tumors. The potential of NK cell-related genes for evaluating the prognosis of patients with hepatocellular carcinoma remains unexplored. This study aims to address this gap by investigating the association between NK cell-related genes and the prognosis of HCC patients, with the goal of developing a reliable model that can provide novel insights into evaluating the immunotherapy response and prognosis of these patients. This work has the potential to significantly advance our understanding of the complex interplay between immune cells and tumors, and may ultimately lead to improved clinical outcomes for HCC patients.
Methods: For this study, we employed transcriptome expression data from the hepatocellular carcinoma cancer genome map (TCGA-LIHC) to develop a model consisting of NK cell-related genes. To construct the NK cell-related signature (NKRLSig), we utilized a combination of univariate COX regression, Area Under Curve (AUC) LASSO COX regression, and multivariate COX regression. To validate the model, we conducted external validation using the GSE14520 cohort.
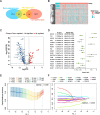
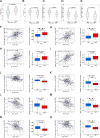
Results: We developed a prognostic model based on 5-NKRLSig (IL18RAP, CHP1, VAMP2, PIC3R1, PRKCD), which divided patients into high- and low-risk groups based on their risk score. The high-risk group was associated with a poor prognosis, and the risk score had good predictive ability across all clinical subgroups. The risk score and stage were found to be independent prognostic indicators for HCC patients when clinical factors were taken into account. We further created a nomogram incorporating the 5-NKRLSig and clinicopathological characteristics, which revealed that patients in the low-risk group had a better prognosis. Moreover, our analysis of immunotherapy and chemotherapy response indicated that patients in the low-risk group were more responsive to immunotherapy.
Conclusion: The model that we developed not only sheds light on the regulatory mechanism of NK cell-related genes in HCC, but also has the potential to advance our understanding of immunotherapy for HCC. With its strong predictive capacity, our model may prove useful in evaluating the prognosis of patients and guiding clinical decision-making for HCC patients.
Keywords: HCC; Natural killer cell; Prognostic signature; Tumor microenvironment.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma[J] Nat Rev Clin Oncol. 2022;19(3):151–172. - PubMed
-
- Vogel A, Meyer T, Sapisochin G, et al. Hepatocellular carcinoma[J] Lancet. 2022;400(10360):1345–1362. - PubMed
-
- Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma[J] Semin Diagn Pathol. 2017;34(2):153–159. - PubMed
-
- Yang JD, Heimbach JK. New advances in the diagnosis and management of hepatocellular carcinoma[J] BMJ. 2020;371:m3544. - PubMed
-
- Tang J, Zhu Q, Li Z, et al. Natural Killer Cell-targeted Immunotherapy for Cancer[J] Curr Stem Cell Res Ther. 2022;17(6):513–526. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

