Live-cell imaging reveals single-cell and population-level infection strategies of Listeria monocytogenes in macrophages
- PMID: 37675103
- PMCID: PMC10478088
- DOI: 10.3389/fimmu.2023.1235675
Live-cell imaging reveals single-cell and population-level infection strategies of Listeria monocytogenes in macrophages
Abstract
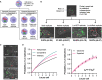
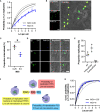
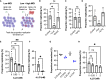
Pathogens have developed intricate strategies to overcome the host's innate immune responses. In this paper we use live-cell microscopy with a single bacterium resolution to follow in real time interactions between the food-borne pathogen L. monocytogenes and host macrophages, a key event controlling the infection in vivo. We demonstrate that infection results in heterogeneous outcomes, with only a subset of bacteria able to establish a replicative invasion of macrophages. The fate of individual bacteria in the same host cell was independent from the host cell and non-cooperative, being independent from co-infecting bacteria. A higher multiplicity of infection resulted in a reduced probability of replication of the overall bacterial population. By use of internalisation assays and conditional probabilities to mathematically describe the two-stage invasion process, we demonstrate that the higher MOI compromises the ability of macrophages to phagocytose bacteria. We found that the rate of phagocytosis is mediated via the secreted Listeriolysin toxin (LLO), while the probability of replication of intracellular bacteria remained constant. Using strains expressing fluorescent reporters to follow transcription of either the LLO-encoding hly or actA genes, we show that replicative bacteria exhibited higher PrfA regulon expression in comparison to those bacteria that did not replicate, however elevated PrfA expression per se was not sufficient to increase the probability of replication. Overall, this demonstrates a new role for the population-level, but not single cell, PrfA-mediated activity to regulate outcomes of host pathogen interactions.
Keywords: Listeria monocytogenes; PrfA regulon; listeriolysin; macrophage; phagocytosis; single cell heterogeneity.
Copyright © 2023 Moran, Feltham, Bagnall, Goldrick, Lord, Nettleton, Spiller, Roberts and Paszek.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

