Use of Epivolve phage display to generate a monoclonal antibody with opsonic activity directed against a subdominant epitope on extracellular loop 4 of Treponema pallidum BamA (TP0326)
- PMID: 37675118
- PMCID: PMC10478084
- DOI: 10.3389/fimmu.2023.1222267
Use of Epivolve phage display to generate a monoclonal antibody with opsonic activity directed against a subdominant epitope on extracellular loop 4 of Treponema pallidum BamA (TP0326)
Abstract
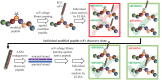
Introduction: Syphilis, a sexually transmitted infection caused by the spirochete Treponema pallidum (Tp), is resurging globally. Tp's repertoire of outer membrane proteins (OMPs) includes BamA (β-barrel assembly machinery subunit A/TP0326), a bipartite protein consisting of a 16-stranded β-barrel with nine extracellular loops (ECLs) and five periplasmic POTRA (polypeptide transport-associated) domains. BamA ECL4 antisera promotes internalization of Tp by rabbit peritoneal macrophages.
Methods: Three overlapping BamA ECL4 peptides and a two-stage, phage display strategy, termed "Epivolve" (for epitope evolution) were employed to generate single-chain variable fragments (scFvs). Additionally, antisera generated by immunizing mice and rabbits with BamA ECL4 displayed by a Pyrococcus furiosus thioredoxin scaffold (PfTrxBamA/ECL4). MAbs and antisera reactivities were evaluated by immunoblotting and ELISA. A comparison of murine and rabbit opsonophagocytosis assays was conducted to evaluate the functional ability of the Abs (e.g., opsonization) and validate the mouse assay. Sera from Tp-infected mice (MSS) and rabbits (IRS) were evaluated for ECL4-specific Abs using PfTrxBamA/ECL4 and overlapping ECL4 peptides in immunoblotting and ELISA assays.
Results: Each of the five mAbs demonstrated reactivity by immunoblotting and ELISA to nanogram amounts of PfTrxBamA/ECL4. One mAb, containing a unique amino acid sequence in both the light and heavy chains, showed activity in the murine opsonophagocytosis assay. Mice and rabbits hyperimmunized with PfTrxBamA/ECL4 produced opsonic antisera that strongly recognized the ECL presented in a heterologous scaffold and overlapping ECL4 peptides, including S2. In contrast, Abs generated during Tp infection of mice and rabbits poorly recognized the peptides, indicating that S2 contains a subdominant epitope.
Discussion: Epivolve produced mAbs target subdominant opsonic epitopes in BamA ECL4, a top syphilis vaccine candidate. The murine opsonophagocytosis assay can serve as an alternative model to investigate the opsonic potential of vaccinogens. Detailed characterization of BamA ECL4-specific Abs provided a means to dissect Ab responses elicited by Tp infection.
Keywords: BamA ECL4; Pyrococcus furiosus thioredoxin; Treponema pallidum; monoclonal antibody; opsonic antibody; outer membrane protein; subdominant epitope; syphilis.
Copyright © 2023 Ferguson, Delgado, McBride, Orbe, La Vake, Caimano, Mendez, Moraes, Schryvers, Moody, Radolf, Weiner and Hawley.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Extracellular Loops of the Treponema pallidum FadL Orthologs TP0856 and TP0858 Elicit IgG Antibodies and IgG+-Specific B-Cells in the Rabbit Model of Experimental Syphilis.mBio. 2022 Aug 30;13(4):e0163922. doi: 10.1128/mbio.01639-22. Epub 2022 Jul 12. mBio. 2022. PMID: 35862766 Free PMC article.
-
A Homology Model Reveals Novel Structural Features and an Immunodominant Surface Loop/Opsonic Target in the Treponema pallidum BamA Ortholog TP_0326.J Bacteriol. 2015 Jun;197(11):1906-20. doi: 10.1128/JB.00086-15. Epub 2015 Mar 30. J Bacteriol. 2015. PMID: 25825429 Free PMC article.
-
Immunodominant extracellular loops of Treponema pallidum FadL outer membrane proteins elicit antibodies with opsonic and growth-inhibitory activities.bioRxiv [Preprint]. 2024 Jul 30:2024.07.30.605823. doi: 10.1101/2024.07.30.605823. bioRxiv. 2024. Update in: PLoS Pathog. 2024 Dec 23;20(12):e1012443. doi: 10.1371/journal.ppat.1012443. PMID: 39131275 Free PMC article. Updated. Preprint.
-
Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Treponema Pallidum Polypeptide Research Group.Microbiol Rev. 1993 Sep;57(3):750-79. doi: 10.1128/mr.57.3.750-779.1993. Microbiol Rev. 1993. PMID: 8246847 Free PMC article. Review.
-
The Treponema pallidum Outer Membrane.Curr Top Microbiol Immunol. 2018;415:1-38. doi: 10.1007/82_2017_44. Curr Top Microbiol Immunol. 2018. PMID: 28849315 Free PMC article. Review.
Cited by
-
Immunodominant extracellular loops of Treponema pallidum FadL outer membrane proteins elicit antibodies with opsonic and growth-inhibitory activities.PLoS Pathog. 2024 Dec 23;20(12):e1012443. doi: 10.1371/journal.ppat.1012443. eCollection 2024 Dec. PLoS Pathog. 2024. PMID: 39715273 Free PMC article.
-
Development and utilization of Treponema pallidum expressing green fluorescent protein to study spirochete-host interactions and antibody-mediated clearance: expanding the toolbox for syphilis research.bioRxiv [Preprint]. 2024 Oct 21:2024.10.21.619476. doi: 10.1101/2024.10.21.619476. bioRxiv. 2024. Update in: mBio. 2025 Jan 8;16(1):e0325324. doi: 10.1128/mbio.03253-24. PMID: 39484466 Free PMC article. Updated. Preprint.
-
T-Cell Responses to Treponema pallidum Proteins in Blood and Skin to Advance Syphilis Vaccine Design: Learning From Nature.J Infect Dis. 2024 Aug 16;230(2):275-277. doi: 10.1093/infdis/jiae246. J Infect Dis. 2024. PMID: 39147388 Free PMC article. No abstract available.
-
Development and utilization of Treponema pallidum expressing green fluorescent protein to study spirochete-host interactions and antibody-mediated clearance: expanding the toolbox for syphilis research.mBio. 2025 Jan 8;16(1):e0325324. doi: 10.1128/mbio.03253-24. Epub 2024 Nov 29. mBio. 2025. PMID: 39611839 Free PMC article.
-
Syphilis vaccine development: Aligning vaccine design with manufacturing requirements.Hum Vaccin Immunother. 2024 Dec 31;20(1):2399915. doi: 10.1080/21645515.2024.2399915. Epub 2024 Sep 11. Hum Vaccin Immunother. 2024. PMID: 39262177 Free PMC article.
References
-
- Radolf JD, Tramont EC, Salazar JC. Syphilis (Treponema pallidum). In: Mandell GL, Dolin R, Blaser MJ, editors. Mandell, Douglas and Bennett's Principles and Practice of Infectious Diseases, 9 ed. Philadelphia: Churchill Livingtone Elsevier; (2019). p. 2865–92.
-
- Radolf JD, Lukehart SA. Immunology of syphilis. In: Radolf JD, Lukehart SA, editors. Pathogenic Treponemes: Cellular and Molecular Biology. Norfolk, UK: Caister Academic Press; (2006). p. 285–322.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

