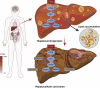
Loss of hepatic FTCD promotes lipid accumulation and hepatocarcinogenesis by upregulating PPARγ and SREBP2
- PMID: 37675273
- PMCID: PMC10477690
- DOI: 10.1016/j.jhepr.2023.100843
Loss of hepatic FTCD promotes lipid accumulation and hepatocarcinogenesis by upregulating PPARγ and SREBP2
Abstract
Background & aims: Exploiting key regulators responsible for hepatocarcinogenesis is of great importance for the prevention and treatment of hepatocellular carcinoma (HCC). However, the key players contributing to hepatocarcinogenesis remain poorly understood. We explored the molecular mechanisms underlying the carcinogenesis and progression of HCC for the development of potential new therapeutic targets.
Methods: The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) and Genotype-Tissue Expression (GTEx) databases were used to identify genes with enhanced expression in the liver associated with HCC progression. A murine liver-specific Ftcd knockout (Ftcd-LKO) model was generated to investigate the role of formimidoyltransferase cyclodeaminase (FTCD) in HCC. Multi-omics analysis of transcriptomics, metabolomics, and proteomics data were applied to further analyse the molecular effects of FTCD expression on hepatocarcinogenesis. Functional and biochemical studies were performed to determine the significance of loss of FTCD expression and the therapeutic potential of Akt inhibitors in FTCD-deficient cancer cells.
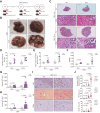
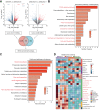
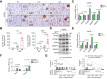
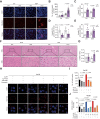
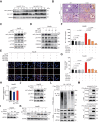
Results: FTCD is highly expressed in the liver but significantly downregulated in HCC. Patients with HCC and low levels of FTCD exhibited worse prognosis, and patients with liver cirrhosis and low FTCD levels exhibited a notable higher probability of developing HCC. Hepatocyte-specific knockout of FTCD promoted both chronic diethylnitrosamine-induced and spontaneous hepatocarcinogenesis in mice. Multi-omics analysis showed that loss of FTCD affected fatty acid and cholesterol metabolism in hepatocarcinogenesis. Mechanistically, loss of FTCD upregulated peroxisome proliferator-activated receptor (PPAR)γ and sterol regulatory element-binding protein 2 (SREBP2) by regulating the PTEN/Akt/mTOR signalling axis, leading to lipid accumulation and hepatocarcinogenesis.
Conclusions: Taken together, we identified a FTCD-regulated lipid metabolic mechanism involving PPARγ and SREBP2 signaling in hepatocarcinogenesis and provide a rationale for therapeutically targeting of HCC driven by downregulation of FTCD.
Impact and implications: Exploiting key molecules responsible for hepatocarcinogenesis is significant for the prevention and treatment of HCC. Herein, we identified formimidoyltransferase cyclodeaminase (FTCD) as the top enhanced gene, which could serve as a predictive and prognostic marker for patients with HCC. We generated and characterised the first Ftcd liver-specific knockout murine model. We found loss of FTCD expression upregulated peroxisome proliferator-activated receptor (PPAR)γ and sterol regulatory element-binding protein 2 (SREBP2) by regulating the PTEN/Akt/mTOR signalling axis, leading to lipid accumulation and hepatocarcinogenesis, and provided a rationale for therapeutic targeting of HCC driven by downregulation of FTCD.
Keywords: Formimidoyltransferase cyclodeaminase; Hepatocarcinogenesis; Lipid metabolism; Tumour suppressor.
© 2023 The Author(s).
Conflict of interest statement
All authors declare no conflicts of interest. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures


Similar articles
-
Formiminotransferase Cyclodeaminase Suppresses Hepatocellular Carcinoma by Modulating Cell Apoptosis, DNA Damage, and Phosphatidylinositol 3-Kinases (PI3K)/Akt Signaling Pathway.Med Sci Monit. 2019 Jun 16;25:4474-4484. doi: 10.12659/MSM.916202. Med Sci Monit. 2019. PMID: 31203308 Free PMC article.
-
Cholesterol biosynthesis supports the growth of hepatocarcinoma lesions depleted of fatty acid synthase in mice and humans.Gut. 2020 Jan;69(1):177-186. doi: 10.1136/gutjnl-2018-317581. Epub 2019 Apr 6. Gut. 2020. PMID: 30954949 Free PMC article.
-
Prognostic alternative splicing and multi-omics characteristics reveal FTCD is a potential target of hepatocellular carcinoma.Discov Oncol. 2024 Nov 27;15(1):718. doi: 10.1007/s12672-024-01201-y. Discov Oncol. 2024. PMID: 39592513 Free PMC article.
-
The ménage à trois of autophagy, lipid droplets and liver disease.Autophagy. 2022 Jan;18(1):50-72. doi: 10.1080/15548627.2021.1895658. Epub 2021 Apr 2. Autophagy. 2022. PMID: 33794741 Free PMC article. Review.
-
PPARγ signaling in hepatocarcinogenesis: Mechanistic insights for cellular reprogramming and therapeutic implications.Pharmacol Ther. 2022 Dec;240:108298. doi: 10.1016/j.pharmthera.2022.108298. Epub 2022 Oct 13. Pharmacol Ther. 2022. PMID: 36243148 Review.
Cited by
-
A novel molecular classification system based on the molecular feature score identifies patients sensitive to immune therapy and target therapy.Front Immunol. 2024 Nov 26;15:1466069. doi: 10.3389/fimmu.2024.1466069. eCollection 2024. Front Immunol. 2024. PMID: 39660131 Free PMC article.
-
Hepatitis B Virus X Protein Upregulates SREBP2 to Modulate Autophagy in Hepatocellular Carcinoma.Cancer Med. 2025 Jun;14(11):e70916. doi: 10.1002/cam4.70916. Cancer Med. 2025. PMID: 40476478 Free PMC article.
-
New insights into T cell metabolism in liver cancer: from mechanism to therapy.Cell Death Discov. 2025 Mar 23;11(1):118. doi: 10.1038/s41420-025-02397-w. Cell Death Discov. 2025. PMID: 40122853 Free PMC article. Review.
-
Novel molecular hepatocellular carcinoma subtypes and RiskScore utilizing apoptosis-related genes.Sci Rep. 2024 Feb 16;14(1):3913. doi: 10.1038/s41598-024-54673-x. Sci Rep. 2024. PMID: 38365931 Free PMC article.
-
Integrative Multi-Omics Approaches Reveal Selectivity Profiles and Molecular Mechanisms of FIIN-2, a Covalent FGFR Inhibitor.Adv Sci (Weinh). 2025 Apr;12(14):e2412578. doi: 10.1002/advs.202412578. Epub 2025 Feb 20. Adv Sci (Weinh). 2025. PMID: 39976135 Free PMC article.
References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. - PubMed
-
- Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. - PubMed
-
- Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–1173. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

