Mitochondrial polymorphism m.3017C>T of SHLP6 relates to heterothermy
- PMID: 37675281
- PMCID: PMC10478271
- DOI: 10.3389/fphys.2023.1207620
Mitochondrial polymorphism m.3017C>T of SHLP6 relates to heterothermy
Abstract
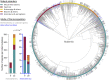
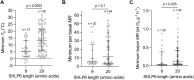
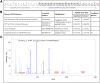
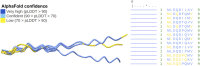
Heterothermic thermoregulation requires intricate regulation of metabolic rate and activation of pro-survival factors. Eliciting these responses and coordinating the necessary energy shifts likely involves retrograde signalling by mitochondrial-derived peptides (MDPs). Members of the group were suggested before to play a role in heterothermic physiology, a key component of hibernation and daily torpor. Here we studied the mitochondrial single-nucleotide polymorphism (SNP) m.3017C>T that resides in the evolutionarily conserved gene MT-SHLP6. The substitution occurring in several mammalian orders causes truncation of SHLP6 peptide size from twenty to nine amino acids. Public mass spectrometric (MS) data of human SHLP6 indicated a canonical size of 20 amino acids, but not the use of alternative translation initiation codons that would expand the peptide. The shorter isoform of SHLP6 was found in heterothermic rodents at higher frequency compared to homeothermic rodents (p < 0.001). In heterothermic mammals it was associated with lower minimal body temperature (T b, p < 0.001). In the thirteen-lined ground squirrel, brown adipose tissue-a key organ required for hibernation, showed dynamic changes of the steady-state transcript level of mt-Shlp6. The level was significantly higher before hibernation and during interbout arousal and lower during torpor and after hibernation. Our finding argues to further explore the mode of action of SHLP6 size isoforms with respect to mammalian thermoregulation and possibly mitochondrial retrograde signalling.
Keywords: SHLP6; daily torpor; extended vertebrate mitochondrial genetic code; hibernation; micropeptide; mitochondrial-derived peptide (MDP); mitogenomics; rodents.
Copyright © 2023 Emser, Spielvogel, Millesi and Steinborn.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
The "minimal boundary curve for endothermy" as a predictor of heterothermy in mammals and birds: a review.J Comp Physiol B. 2008 Jan;178(1):1-8. doi: 10.1007/s00360-007-0193-0. Epub 2007 Aug 3. J Comp Physiol B. 2008. PMID: 17674009 Review.
-
Seasonal changes in brown adipose tissue mitochondria in a mammalian hibernator: from gene expression to function.Am J Physiol Regul Integr Comp Physiol. 2016 Aug 1;311(2):R325-36. doi: 10.1152/ajpregu.00463.2015. Epub 2016 May 25. Am J Physiol Regul Integr Comp Physiol. 2016. PMID: 27225952
-
Extensive use of torpor in 13-lined ground squirrels in the fall prior to cold exposure.J Comp Physiol B. 2010 Nov;180(8):1165-72. doi: 10.1007/s00360-010-0484-8. Epub 2010 Jun 17. J Comp Physiol B. 2010. PMID: 20556614 Free PMC article.
-
Evolution of daily torpor and hibernation in birds and mammals: importance of body size.Clin Exp Pharmacol Physiol. 1998 Sep;25(9):736-9. doi: 10.1111/j.1440-1681.1998.tb02287.x. Clin Exp Pharmacol Physiol. 1998. PMID: 9750966 Review.
-
Arousal from Torpor Increases Oxidative Damage in the Hibernating Thirteen-Lined Ground Squirrel (Ictidomys tridecemlineatus).Physiol Biochem Zool. 2022 May-Jun;95(3):229-238. doi: 10.1086/719931. Physiol Biochem Zool. 2022. PMID: 35443147
Cited by
-
Mitochondrial-derived gene expression in hibernation: tissue-specific responses in the thirteen-lined ground squirrel.Open Biol. 2025 Aug;15(8):240255. doi: 10.1098/rsob.240255. Epub 2025 Aug 13. Open Biol. 2025. PMID: 40795995 Free PMC article.
References
LinkOut - more resources
Full Text Sources

