The role of SARS-CoV-2 N protein in diagnosis and vaccination in the context of emerging variants: present status and prospects
- PMID: 37675423
- PMCID: PMC10478715
- DOI: 10.3389/fmicb.2023.1217567
The role of SARS-CoV-2 N protein in diagnosis and vaccination in the context of emerging variants: present status and prospects
Abstract
Despite many countries rapidly revising their strategies to prevent contagions, the number of people infected with Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to surge. The emergent variants that can evade the immune response significantly affect the effectiveness of mainstream vaccines and diagnostic products based on the original spike protein. Therefore, it is essential to focus on the highly conserved nature of the nucleocapsid protein as a potential target in the field of vaccines and diagnostics. In this regard, our review initially discusses the structure, function, and mechanism of action of N protein. Based on this discussion, we summarize the relevant research on the in-depth development and application of diagnostic methods and vaccines based on N protein, such as serology and nucleic acid detection. Such valuable information can aid in designing more efficient diagnostic and vaccine tools that could help end the SARS-CoV-2 pandemic.
Keywords: SARS-CoV-2; diagnostic methods; immune escape; nucleocapsid protein; vaccine development; variants of concern.
Copyright © 2023 Song, Fang, Ma, Li, Huang, Zhang, Li and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
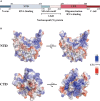
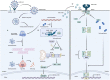



References
-
- Alkhatib M., Bellocchi M., Marchegiani G., Grelli S., Micheli V., Stella D., et al. (2022). First case of a COVID-19 patient infected by delta AY.4 with a rare deletion leading to a N gene target failure by a specific real time PCR assay: Novel omicron VOC might be doing similar scenario? Microorganisms 10:268. 10.3390/microorganisms10020268 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

