MV-CVIB: a microbiome-based multi-view convolutional variational information bottleneck for predicting metastatic colorectal cancer
- PMID: 37675425
- PMCID: PMC10477591
- DOI: 10.3389/fmicb.2023.1238199
MV-CVIB: a microbiome-based multi-view convolutional variational information bottleneck for predicting metastatic colorectal cancer
Abstract
Introduction: Imbalances in gut microbes have been implied in many human diseases, including colorectal cancer (CRC), inflammatory bowel disease, type 2 diabetes, obesity, autism, and Alzheimer's disease. Compared with other human diseases, CRC is a gastrointestinal malignancy with high mortality and a high probability of metastasis. However, current studies mainly focus on the prediction of colorectal cancer while neglecting the more serious malignancy of metastatic colorectal cancer (mCRC). In addition, high dimensionality and small samples lead to the complexity of gut microbial data, which increases the difficulty of traditional machine learning models.
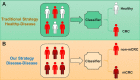
Methods: To address these challenges, we collected and processed 16S rRNA data and calculated abundance data from patients with non-metastatic colorectal cancer (non-mCRC) and mCRC. Different from the traditional health-disease classification strategy, we adopted a novel disease-disease classification strategy and proposed a microbiome-based multi-view convolutional variational information bottleneck (MV-CVIB).
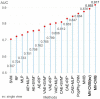
Results: The experimental results show that MV-CVIB can effectively predict mCRC. This model can achieve AUC values above 0.9 compared to other state-of-the-art models. Not only that, MV-CVIB also achieved satisfactory predictive performance on multiple published CRC gut microbiome datasets.
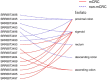
Discussion: Finally, multiple gut microbiota analyses were used to elucidate communities and differences between mCRC and non-mCRC, and the metastatic properties of CRC were assessed by patient age and microbiota expression.
Keywords: information bottleneck; metastatic colorectal cancer; microbiome; multi-view; risk assessment.
Copyright © 2023 Cui, Wu, Zhang, Wang, He and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Using gut microbiota as a diagnostic tool for colorectal cancer: machine learning techniques reveal promising results.J Med Microbiol. 2023 Jun;72(6). doi: 10.1099/jmm.0.001699. J Med Microbiol. 2023. PMID: 37288545
-
A Goldilocks Principle for the Gut Microbiome: Taxonomic Resolution Matters for Microbiome-Based Classification of Colorectal Cancer.mBio. 2022 Feb 22;13(1):e0316121. doi: 10.1128/mbio.03161-21. Epub 2022 Jan 11. mBio. 2022. PMID: 35012354 Free PMC article.
-
A multiple-dimension model for microbiota of patients with colorectal cancer from normal participants and other intestinal disorders.Appl Microbiol Biotechnol. 2022 Mar;106(5-6):2161-2173. doi: 10.1007/s00253-022-11846-w. Epub 2022 Feb 26. Appl Microbiol Biotechnol. 2022. PMID: 35218389
-
Dietary Efficacy Evaluation by Applying a Prediction Model Using Clinical Fecal Microbiome Data of Colorectal Disease to a Controlled Animal Model from an Obesity Perspective.Microorganisms. 2022 Sep 14;10(9):1833. doi: 10.3390/microorganisms10091833. Microorganisms. 2022. PMID: 36144434 Free PMC article. Review.
-
Gut Microbiome in Colorectal Cancer: Clinical Diagnosis and Treatment.Genomics Proteomics Bioinformatics. 2023 Feb;21(1):84-96. doi: 10.1016/j.gpb.2022.07.002. Epub 2022 Jul 30. Genomics Proteomics Bioinformatics. 2023. PMID: 35914737 Free PMC article. Review.
References
-
- Alemi A. A., Fischer I., Dillon J. V., Murphy K. (2016). Deep variational information bottleneck. arXiv. [preprint]. 10.48550/arXiv.1612.00410 - DOI
LinkOut - more resources
Full Text Sources

